


Did you find this useful? Give us your feedback





128 citations
52 citations
38 citations
11 citations
10 citations
27,771 citations
22,622 citations
16,857 citations
...…genomic analyses is widely recognized for estimating dates of127 emergence(Verity Hill, 2020; Gire et al., 2014) and identifying animal reservoirs(Zhou et al., 2020;128 Dudas et al., 2018), analysis of pathogen sequences also has potential to inform epidemic surveil-129 lance and intervention…...
[...]
4,337 citations
...A time scaled phylogeny estimated using IQTree and treedater and using the same data as used for the Bayesian analysis....
[...]
...Maximum likelihood analysis was carried using IQTree(Minh et al., 2019) with a HKY+G4173 substitution model and a time-scaled tree was estimated using treedater 0.5.0(Volz and Frost, 2017).174 Two outliers according to the molecular clock model were identified and removed using ‘treedater’175 which was also used to compute the root to tip regression.176 Bayesian phylogenetic analysis was carried out using BEAST 2.6.1(Bouckaert et al., 2019) using a177 HKY+G4 substitutionmodel and a strict molecular clock....
[...]
...Maximum likelihood analysis was carried using IQTree(Minh et al., 2019) with a HKY+G4173 substitution model and a time-scaled tree was estimated using treedater 0.5.0(Volz and Frost, 2017).174 Two outliers according to the molecular clock model were identified and removed using ‘treedater’175 which…...
[...]
3,467 citations
...We use a susceptible-exposed-infectious-recovered (SEIR) model(Keeling and Rohani, 2011) 58 for epidemic dynamics in Weifang....
[...]
...We use a susceptible-exposed-infectious-recovered (SEIR) model(Keeling and Rohani, 2011)58 for epidemic dynamics in Weifang....
[...]
The phylodynamic model is designed to account for 1, nonlinear epidemic dynamics in Weifang with a realistic course of infection (incubation and infectious periods), 2, variance in transmission rates that can influence epidemic size estimates, and 3, migration of lineages in and out of Weifang.
The Coronaviridae-like reads of samples with >100 average sequencing depth across the SARS-CoV-2 genome were subsampled to achieve 100 sequencing depth before being assembled.
Although other methods which allow for time-varying transmission rate (including other PhyDyn model templates) or models with a piece-wise Rt function (Frost and Volz 2010), their SEIR-type model with constant b required fewer parameters, appropriate for an analysis with only 20 internal sequences.
Total reads were first processed using Kraken v0.10.5 (default parameters) with a self-built database of Coronaviridae genomes (including SARS, MERS, and SARS-CoV-2 genome sequences downloaded from GISAID, NCBI, and CNGB) to identify Coronaviridae-like reads.
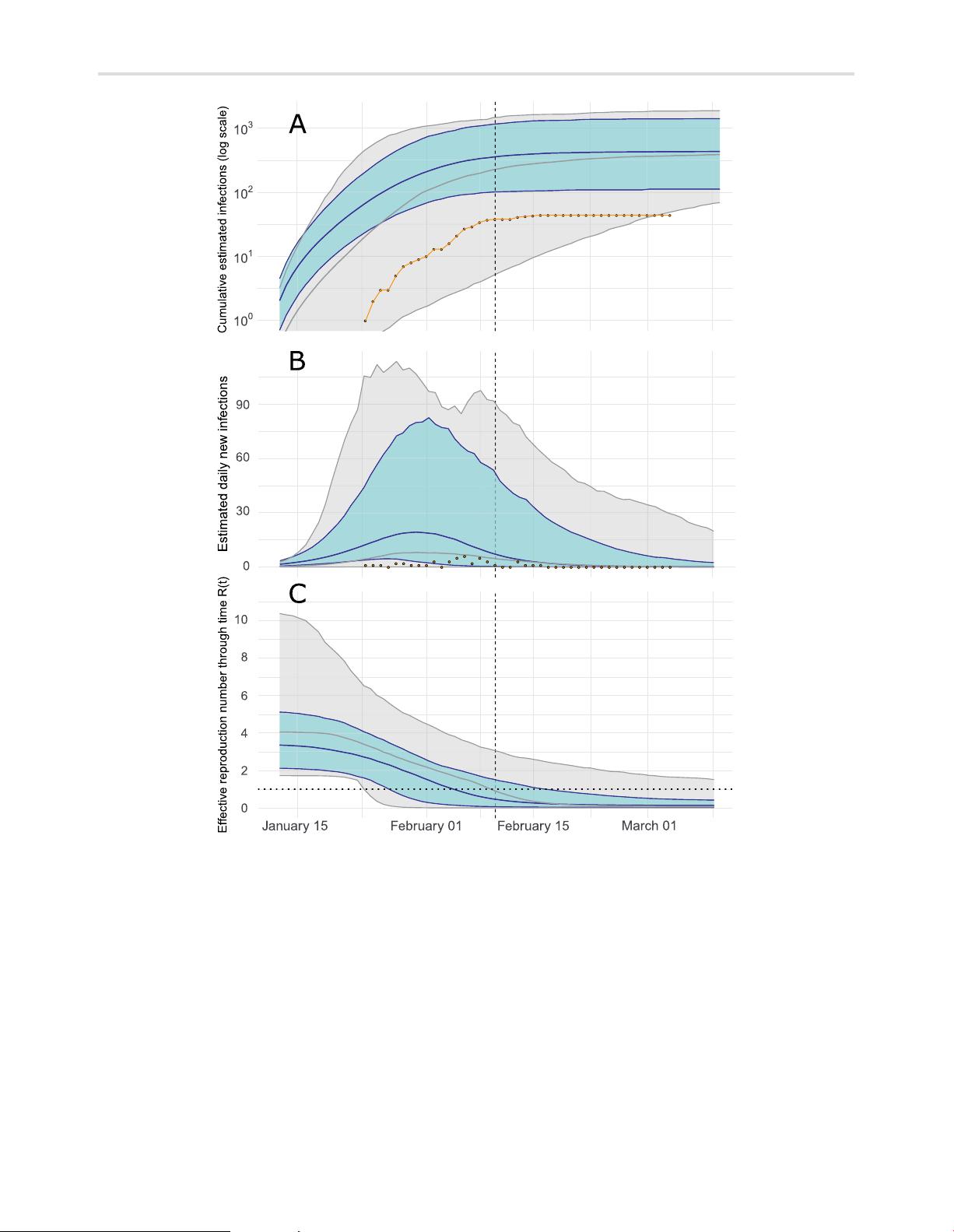
The authors estimate the peak of daily infections in late January, preceding the time series of confirmed cases by about a week; this is expected due to delays from infection to appearance of symptoms and delays from symptoms to diagnosis.
These interventions included public health messaging, establishing phone hot-lines, encouraging home isolation for recent visitors from Wuhan (January 23–26), optimising triage of suspected cases in hospitals (January 24), travel restrictions (January 26), extending school closures, and establishing ‘fever clinics’ for consultation and diagnosis (January 27) (Mao 2020).
The larger reservoir of COVID-19 cases outside of Weifang (Y (t)) serves as a source of new infections and is assumed to be growing exponentially (at rate q) over this time period.
the added value of fitting to only 20 local sequences in this analysis demonstrates the utility of phylodynamic modelling for outbreaks as compared with traditional epidemiological modelling fitted only to case data.
As of 10 February 2020, 136 suspected cases and 214 close contacts were diagnosed by Weifang Center for Disease Control and Prevention; of these, 38 cases were confirmed positive with SARS-CoV-2.
Im perial C ollege London Library user on 29 M arch 2021thirty-eight confirmed cases at the date of the last genetic sample (10 February), rising no further than forty-four from 16 February onwards (Fig. 2A).
High variance of transmission rates will reduce genetic diversity of a sample and failure to account for this factor will lead to highly biased estimates of epidemic size (Li et al. 2017).
The remaining RNA was used to construct the single-stranded circular DNA library with the MGIEasy RNA Library preparation reagent set (MGI, Shenzhen, China).
This is likely due to their choice of reference sequence set, which comprised sequences spanning several months of the epidemic, and therefore reflecting a range of transmission dynamics.