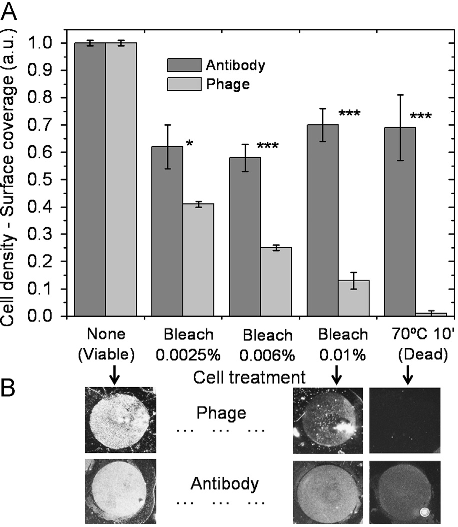
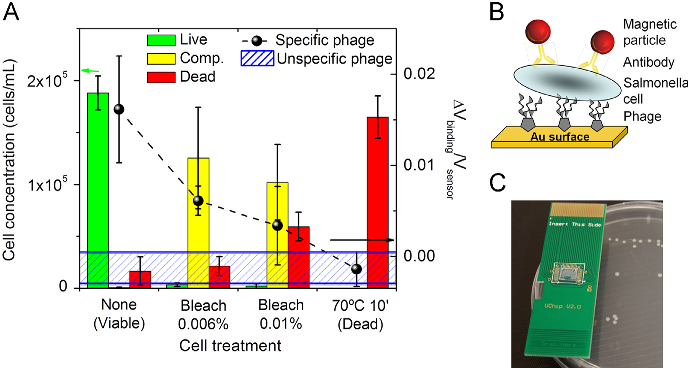
Abstract: Molecular Cloning has served as the foundation of technical expertise in labs worldwide for 30 years. No other manual has been so popular, or so influential. Molecular Cloning, Fourth Edition, by the celebrated founding author Joe Sambrook and new co-author, the distinguished HHMI investigator Michael Green, preserves the highly praised detail and clarity of previous editions and includes specific chapters and protocols commissioned for the book from expert practitioners at Yale, U Mass, Rockefeller University, Texas Tech, Cold Spring Harbor Laboratory, Washington University, and other leading institutions. The theoretical and historical underpinnings of techniques are prominent features of the presentation throughout, information that does much to help trouble-shoot experimental problems. For the fourth edition of this classic work, the content has been entirely recast to include nucleic-acid based methods selected as the most widely used and valuable in molecular and cellular biology laboratories. Core chapters from the third edition have been revised to feature current strategies and approaches to the preparation and cloning of nucleic acids, gene transfer, and expression analysis. They are augmented by 12 new chapters which show how DNA, RNA, and proteins should be prepared, evaluated, and manipulated, and how data generation and analysis can be handled. The new content includes methods for studying interactions between cellular components, such as microarrays, next-generation sequencing technologies, RNA interference, and epigenetic analysis using DNA methylation techniques and chromatin immunoprecipitation. To make sense of the wealth of data produced by these techniques, a bioinformatics chapter describes the use of analytical tools for comparing sequences of genes and proteins and identifying common expression patterns among sets of genes. Building on thirty years of trust, reliability, and authority, the fourth edition of Mol