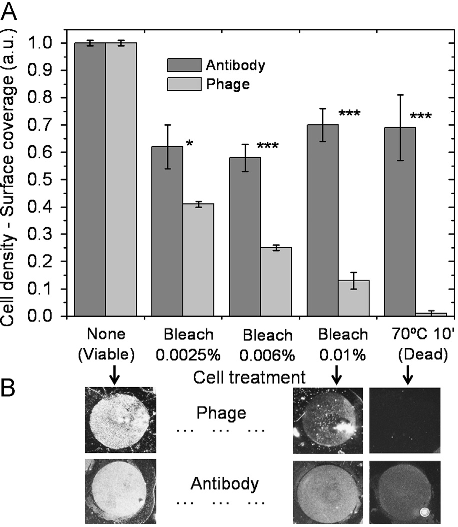
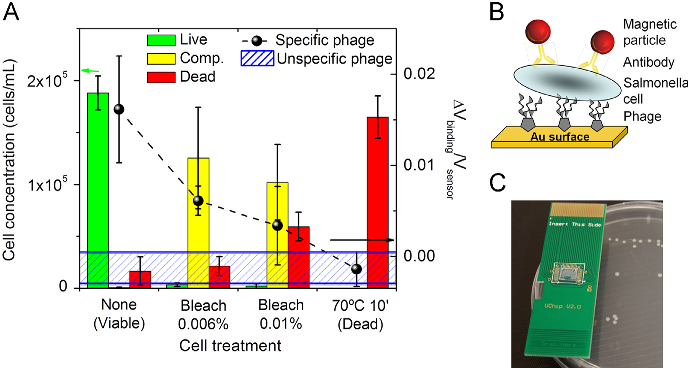
Abstract: Despite the recent advances in food pathogen detection, there still exist many challenges and opportunities to improve the current technology. Techniques such as immunomagnetic separation (IMS) and polymerase chain reaction (PCR) have paved the way for rapid and sensitive detection of foodborne pathogens, and advances in nanobiotechnology have allowed for miniaturization of devices. Collaborations between workers in the fields of engineering, nanotechnology and food science have introduced new lab-on-a-chip technologies permitting development of portable, hand-held biosensors for food pathogen detection. This report highlights examples within the current state of the art, and emphasizes areas in which further research is needed.
In 1999 it was estimated that foodborne pathogens were responsible for 76 million illnesses annually, resulting in 5,000 deaths [1]. This report identified Salmonella, Listeria and Toxoplasma as the major causative agents, being responsible for 1,500 of the reported deaths. Data published in 2006 by the CDC suggested that infections due to Yersinia, Shigella, Listeria, Campylobacter, Escherichia coli O157:H7 and Salmonella have decreased dramatically, while infections due to Vibrio have increased [2]. A more recent report indicated similar findings, with a decrease in Yersinia, Shigella, Listeria and Campylobacter cases, and again a significant increase in Vibrio infections [3]. The declining rates of infection due to Listeria monocytogenes and E. coli O157:H7 are likely a result of increased awareness. The FDA, USDA and EU have all implemented a zero-tolerance rule for L. monocytogenes in ready-to-eat (RTE) foods. Similarly, the USDA’s Food Safety and Inspection Service (USDA-FSIS) has declared E. coli O157:H7 in raw ground beef to be an adulterant and therefore unfit for human consumption. Food safety practices have vastly improved in the processing environment as a result of these regulatory actions, as evidenced by the decreasing rates of infection by both Listeria and E. coli O157:H7. However, of the 121 foodborne outbreaks reported through FoodNet in 2005, almost half (49%) of the reported cases were attributed to noroviruses, and the number of Vibrio infections is on the rise [2]. Therefore, while the “hot” organism may change and while food safety practices are improving, there remains a growing need for enhanced means of food pathogen detection.
In addition to the health risk associated with contaminated foods, there is the often devastating economic impact to the food producer. A 2007 recall of 21. 7 × 106 lb of ground beef owing to contamination with E. coli O157:H7 resulted in the Topps Meat Company going out of business after 67 years of operation. Indeed, the consolidation of food producers in the USA means that the larger companies have a greater responsibility toward protecting the food supply. Coast-to-coast and international distribution by these mega-processing plants puts potential outbreaks on a national and international scale. Increasing automation in food processing facilities increases the risk of contamination by environmental sources following heat treatment, which is a critical concern especially for RTE products. Therefore, monitoring of pathogen counts on processing surfaces is critical in maintaining low or zero counts in food products.
The costs of warehousing along with the potential costs of product recalls have potentially made on-site pathogen testing economically advantageous. Faster results would mean that products could go to market earlier. Several companies such as Marshfield Food Safety and IEH now offer on-site testing services for food production facilities. The latter can equip a production facility with modular laboratories that can be brought in, providing faster turnaround results. These needs stem from the increasing number of microbiological tests commissioned for analysis each week. A 2000 study estimated the number of weekly tests commissioned per dairy plant to be 636, followed closely by 444 weekly microbiological tests from processed-food plants [4].