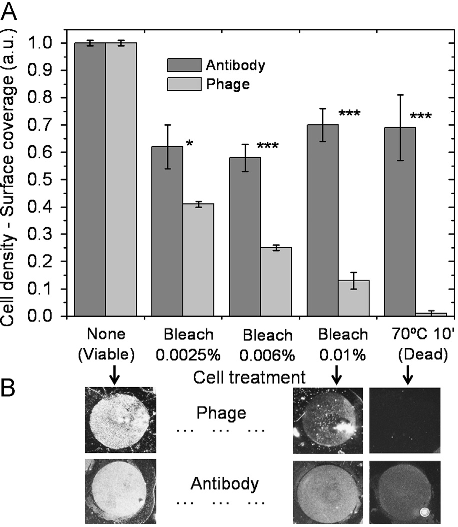
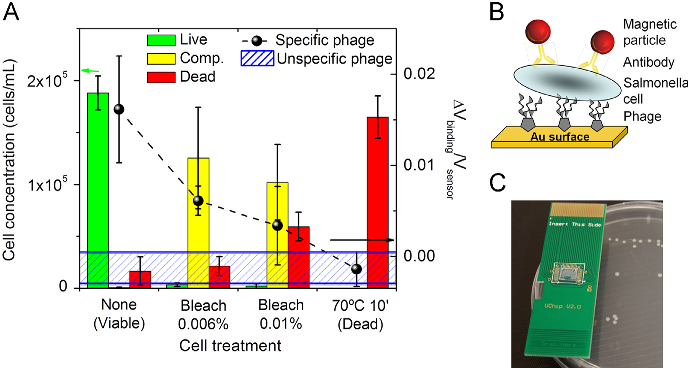
Abstract: BACKGROUND: Conventional techniques used to assess bactericidal activities of antibodies are time-consuming; flow cytometry has been used as a rapid alternative. In this study, the membrane potential-sensitive fluorescent probes bis-(1,3-dibutylbarbituric acid) trimethine oxonol (DiBAC4(3)) and Sytox Green, the redox dye cyano-2,3-ditolyl tetrazolium chloride (CTC), and the Baclite viability test kit were used to assess the effects of ceftazidime, ampicillin, and vancomycin on clinical isolates of Pseudomonas aeruginosa, Escherichia coli, and Staphylococcus aureus, respectively. METHODS: Bacterial cultures were grown to early exponential phase, at which point the antibiotics were added at their breakpoint values, and incubation was allowed to continue. At timed intervals, samples were stained and flow cytometric analysis was performed on a Skatron Argus 100 arc-lamp based dual-parameter flow cytometer. RESULTS: All the dyes successfully identified antibiotic-induced damage in the three strains, although different fluorescence responses between the dyes were observed. DiBAC4(3) and Sytox Green overestimated numbers of nonviable bacteria relative to loss of viability as judged by plate counts. CTC, a measure of respiratory activity, revealed antibiotic-induced population heterogeneity illus trated by the development of several subpopulations. The "live" component of the viability kit identified two populations corresponding to viable and nonviable organisms, whereas the "dead" component only revealed single populations, the fluorescence intensity of which increased with antibiotic exposure. CONCLUSIONS: Flow cytometry provides a rapid and sensitive technique for the evaluation of the antibacterial activities of antibiotics. The use of a range of fluorophores specific for different cellular characteristics may be beneficial, bearing in mind the different fluorescence responses observed among the dyes used here.