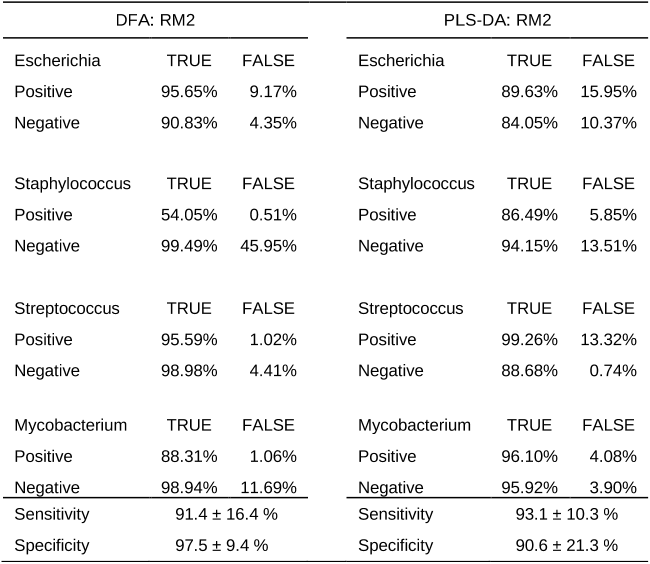
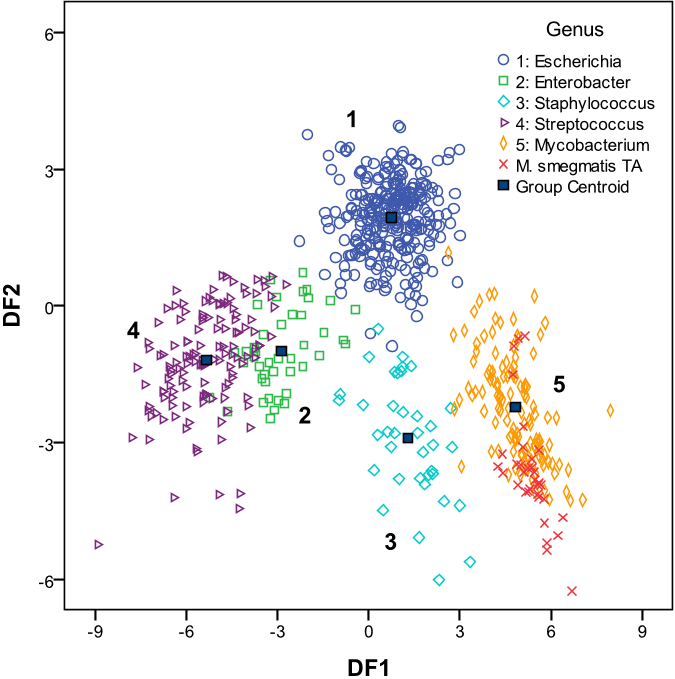
Abstract: En esta tesis se ha estudiado el enfoque cualitativo de la tecnica LIBS en el area de la biomedicina, explorando su potencial para identificar y discriminar muestras biologicas, a partir de los cambios en su composicion atomica. La motivacion de estos estudios fue evaluar la capacidad de LIBS para proporcionar una identificacion rapida en comparacion con los metodos bioanaliticos tradicionales, aprovechando la posibilidad de combinarlo con los metodos quimiometricos para aumentar el rendimiento de la tecnica y demostrar su potencial de uso como un metodo diagnostico en el ambito clinico. Con el fin de desarrollar los modelos de clasificacion, se emplearon diferentes enfoques de los metodos quimiometricos y se compararon para encontrar el mejor enfoque y dar solucion a este problema. Una parte integral es el desarrollo de modelos de clasificacion utilizando los algoritmos de Red Neural Artificial (NN) como la herramienta quimiometrica para el analisis de datos espectrales LIBS en la identificacion y discriminacion de materiales biologicos moleculares complejos como bacterias y hongos. La seleccion de la NN fue fomentada por un estudio en el que la comparacion entre varios metodos quimiometricos, incluyendo Analisis Discriminante Lineal (LDA), Maquinas de Soporte Vectorial (SVM), Modelado Suave Independiente de Analogia de Clase (SIMCA), Analisis Discrecional de minimos cuadrados parciales (PLSDA) y Redes Neuronales Artificiales (NN), demostro el mejor desempeno de la NN en la clasificacion de cepas bacterianas. Hay una multitud de tecnicas que se emplean para identificar los patogenos bacterianos y fungicos presentes en una muestra biologica que causa enfermedades infecciosas. Todas estas tecnicas ofrecen varias ventajas y proporcionan buenos resultados, pero en muchos casos los factores como el tiempo y el coste del analisis son limitados. Por lo tanto, en esta tesis se pretendio desarrollar una metodologia basada en la combinacion LIBS con NN para realizar la identificacion y discriminacion de estos patogenos en la identificacion y discriminacion de bacterias y hongos en muestras biologicas; con especial referencia a la mejora de los estandares de seguridad hospitalaria, particularmente desde el punto de vista microbiologico, mediante el diagnostico precoz de las infecciones adquiridas en el hospital (HAI). La tesis se divide principalmente en tres partes: Introduccion a los fundamentos de las tecnicas utilizadas, procedimiento Experimental y Resultados. La primera parte se centra en la introduccion al objetivo principal de esta tesis. El capitulo 1 da una vision teorica de la tecnica incluyendo los fundamentos de la tecnica LIBS, los antecedentes del analisis basado en LIBS de microorganismos y los avances realizados en esta tecnica para sus aplicaciones medicas. El capitulo 2 incluye una introduccion general a la quimiometria y las figuras de merito que se necesitan tener en cuenta para la evaluacion del desempeno de los metodos quimiometricos en clasificacion. La segunda parte que compone el capitulo 3 describe la configuracion experimental utilizada para las mediciones LIBS y los componentes del sistema experimental desarrollado en el laboratorio. La tercera parte incluye los resultados experimentales donde el capitulo 4 trata sobre la metodologia desarrollada de LIBS y NN para la identificacion y discriminacion de muestras bacterianas. La metodologia se extendio a cepas bacterianas resistentes a antibioticos, que forma la segunda subseccion. Tambien se realizo una comparacion entre diferentes metodos quimiometricos, formando la ultima parte de este capitulo. El capitulo 5 presenta el analisis de muestras de hongos (Candida) por LIBS, su caracterizacion por SEM con EDS, y aplicando la metodologia LIBS y NN para su discriminacion. Finalmente, la ultima parte, el capitulo 6 discute las conclusiones del trabajo y los desafios que enfrentan en este campo.