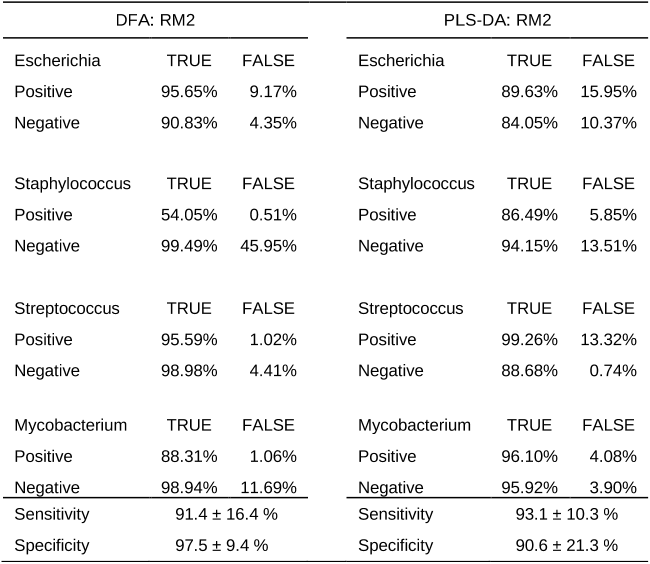
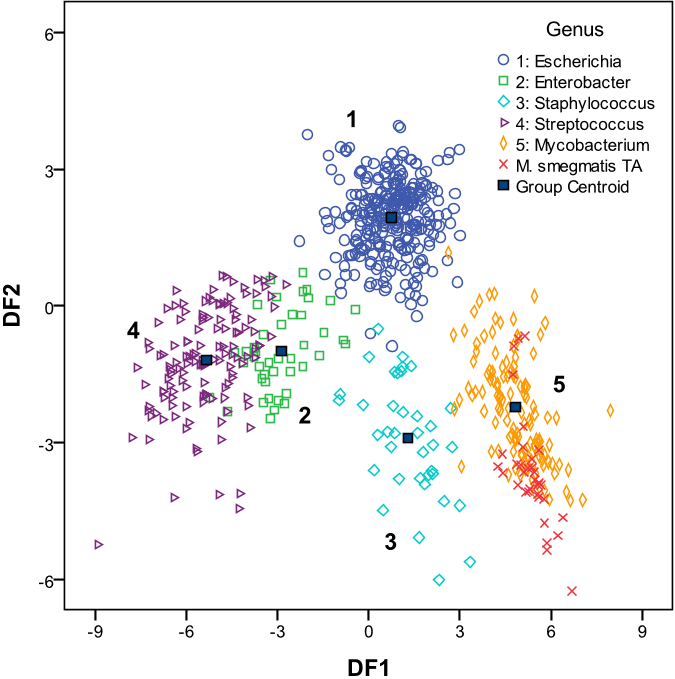
Abstract: Recent advances in high-throughput methods of molecular analyses have led to an explosion of studies generating large-scale ecological data sets. In particular, noticeable effect has been attained in the field of microbial ecology, where new experimental approaches provided in-depth assessments of the composition, functions and dynamic changes of complex microbial communities. Because even a single high-throughput experiment produces large amount of data, powerful statistical techniques of multivariate analysis are well suited to analyse and interpret these data sets. Many different multivariate techniques are available, and often it is not clear which method should be applied to a particular data set. In this review, we describe and compare the most widely used multivariate statistical techniques including exploratory, interpretive and discriminatory procedures. We consider several important limitations and assumptions of these methods, and we present examples of how these approaches have been utilized in recent studies to provide insight into the ecology of the microbial world. Finally, we offer suggestions for the selection of appropriate methods based on the research question and data set structure.