A fast and reliable strategy to generate TALEN-mediated gene knockouts
in the diatom Phaeodactylum tricornutum
M. Serif
1
, B. Lepetit
1
, K. Weißert, P.G. Kroth, C. Rio Bartulos
⁎
Plant Ecophysiology, Fachbereich Biologie, Universität Konstanz, D-78457 Konstanz, Germany
abstract
Reverse genetics techniques are powerful tools for studying gene functions. In the model diatom Phaeodactylum
tricornutum, RNAi-mediated knockdown of genes still is the most commonly used reverse genetics technique.
Due to the diploidic life cycle missing reproduction in lab cultures, many commonly used techniques to create
knockout instead of knockdown lines are not applicable in P. tricornutum. These limitations can be overcome
by using genome editing approaches like TALEN (Transcription activator-like effector nucleases), and/or
CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats), allowing the introduction of targeted
mutagenesis events. Both techniques have recently been adapted exemplarily for diatoms, however, no concise
guidelines exist yet for routine utilization of these tools and the subsequent characterization of the mutants. We
therefore have adapted a cost-effective TALEN generation system previously established for mammalian cells for
the use in P. tricornutum, allowing the assembly of TALENs in about two weeks. We further provide protocols for:
a) choosing a TALEN target site in order to avoid potentially ineffective and/or off-target prone TALEN constructs,
b) efficient transformation of P. tricornutum with both TALEN constructs, utilizi ng two antibiotics resistance
markers, c) effective screening of the transformants. In order to test our system we chose the blue-light depen-
dent transcription factor Aureochrome 1a (PtAureo1a) as a target gene due to the known phenotype of previously
characterized P. tricornutu m RNAi knockdown strains. Our TALEN approach appears to be highly efficient:
targeted mutation events were detected in 50% of all transformants obtained, whereas 21% of the transformants
were found to be bi-allelic knockout lines. Furthermore, most TALEN transformed cell lines were found to be ge-
netically homogeneous without the need for re-plating, which greatly facilitates the screening process.
.
Keywords:
Phaeodactylum tricornutum
TALEN
PtAUREO1a knockout
Blue light-dependent transcription factor
1. Introduction
Diatoms are unicellular microalgae belonging to the Stramenopiles.
They play an important role in global carbon fixation as well as for the
nitrogen, phosphorous and silica cycles [1]. They are widespread in
most aquatic habitats, where they need to cope with large variations
of light quality and quantity [2,3]. As diatoms may contain larger
amounts of lipids (up to 50% of dry weight), which even can be in-
creased by genetic manipulation [4,5], they are suitable for the produc-
tion of biodiesel and/or bioplastics [6–8]. The pennate Phaeodactylum
tricornutum has becom e a model system fo r diatoms because of the
availability of the genome sequence as well as genetic transformation
techniques [9–14], allowing reverse genetics approaches. Because of
their diplontic life cycle and the lack of of sexual reproduction in the
lab, many methods for genetic manipulation like random mutagenesis
or crossing are not available. Accordingly, knockdown via RNAi is
currently the most commonly used approach for genetic manipulations
[15]. Recently, two new genetic tools for directed genome editing were
developed, whi ch allow induction of targeted DNA double-strand
breaks to knock out genes irreversibly: the TALEN (Transcription
activator-like effector nucleases) [16–18] and the CRISPR/Cas9 (clus-
tered regularly interspaced short palindromic repeats) systems
[19–21]. Both approaches depend on nucleases that are guided to a spe-
cific DNA target sequence, and subsequently induce the formation of a
DNA double strand break. The sequence-specific DNA bindin g of
TALEN proteins is based on multiple 34 amino acid repeat modules,
termed repeat variable di-residues (RVD), each binding specifically to
one of the four nucleotides. All modules together (termed “targeting se-
quence”) allow the recognition of a specific DNA sequence, so that in
principle any DNA sequence of interes t within the genome can be
targeted [22,23]. The catalytic domain of the endonuclease FokI, being
fused to this targeting sequence, is only active as a dimer. Hence, for suc-
cessful FokI activity two TALEN proteins are required to bind in the right
orientation and in close proximity onto the DNA double strand to induce
double strand break formation. The mandatory binding of both TALENs
strongly increases the targeting specificity [24]. The double strand break
⁎ Corresponding author.
E-mail address: riobartulos@gmail.com (C. Rio Bartulos).
1
both authors contributed equally.
Konstanzer Online-Publikations-System (KOPS)
URL: http://nbn-resolving.de/urn:nbn:de:bsz:352-2-6ss0tqyo960o9
Erschienen in: Algal Research ; 23 (2017). - S. 186-195.
https://dx.doi.org/10.1016/j.algal.2017.02.005

can be subsequently repaired by cellular mechanisms based either on
homologous recombination (HR) or on non-homologous end joining
(NHEJ). While NHEJ occurs during the whole cell cycle, HR is mainly re-
stricted to the late S and G2 phase [25]. Both of these DNA repair mech-
anisms can be used to induce targeted mutations: HR can be exploited
by introduction of foreign DNA with a strong homology to the DNA se-
quence surrounding the target site, which the cell uses as template to
repair the double strand break [26]. If no HR template is used, the high
error rate of NHEJ can be ex ploited to generate small random
insertions/deletions until the target site is inaccessible for TALEN [27].
The successful application of TALEN and CRISPR/Cas9 in
P. tricornutum has been published recently [28–30], however, a number
of potential pitfalls have not yet been sufficiently addressed. Therefore,
in this report, we describe how the TALEN approach can be optimized to
obtain cell lines with targeted mutations at a high frequency and how to
minimize the risk of potential off-target binding of TALENs. Additionally,
we show that a thorough screening process is required for correctly
distinguishing bi-allelic and mono-allelic knockout mutants, as well as
to prove that a specific cell line is genetically homogeneous and does
not contain different genotype lines. Although the CRISPR approach
can be more easily adapted for a specific target site, we chose to improve
the strategy of utilization of TALENs in diatoms because, base d on
research in other organis ms [31], it offers the potential for reduced
off-target eff ects. We developed a diatom-specific TALEN system by
combining two previously published P. tricornutum transformation
plasmids, pPha-T1 (Genebank ID: AF219942.1, [13]) and pPha-NR
(Genebank ID: JN180663.1, [32]), and the TALEN plasmids developed
by the Zhang lab for mammalian systems [33]. We adapted the Zhang
protocol [33] for TALENs assembly and verification for diatoms, allowing
the creation of TALENs in less than two weeks. We furthermore devel-
oped concise guidelines for all steps of this method, from target site de-
sign to screening mutated lines, which allows any molecular biology lab
equipped with a particle gun or an electroporator to produce knockout
mutants of P. tricornutum. In order to evaluate our TALEN system, we
generated TALENs targeting the PtAureo1a gene, encoding the blue-
light photoreceptor Aureochrome 1a (PtAUREO1a), because of the
availability of a specific antiserum as well as the known phenotype of
RNAi-silenced strains, such as lower chlorophyll a (Chl a) content per
cell and a higher relative amount of xanthophyll cycle pigments [35].
2. Material & methods
2.1. Assembly of the TALEN plasmids
The plasmid kit used for building TALENs was a gift from Dr. Feng
Zhang (Addgene, Cambridge, MA, USA; kit #1000000019) [33].AHindIII
restriction site was introduced upstream of the start codon of the TALEN
plasmids using primer pair TALEN_HindIII + _for/rev (see Table SI). The
P. tricornutum expression plasmids pPha-T1 (GenBank AF219942, [13])
and pPha-NR (GenBank JN180663, [32])weremodifi
ed u sing site-
directe
d
mutagenesis with the primer pairs PTV_BSAI1719SD_for/rev
and PTV_BSAI2888SD_for/rev (see Table SI) to remove the two BsaI re-
striction sites. Furthermore, the Zeocin resistance gene (Sh ble) from the
pPha-NR vector was exchanged with the Nourseothricin resistance gene
(nat) gene from the pNat vector [13], creating the pPha-NR-Nat
vector. The different backbones of the TALEN expression plasmids
from [33] (pTALEN_v2_NG (de tects T ), pTALEN_v2_NI (detects A),
pTALEN_v2_NN (detects G or A), pTALEN_v2_HD (detects C)) were ex-
cised from the respective plasmids using a HindIII/SacI double digest,
and ligated into both pPha T1 and pPha-NR-Nat to create
P. tricornutum specific TALEN plasmids pM9_fc pA_NG, pM9_fcpA_NI,
pM9_fcpA_NN, pM9_fcpA_HD and pM9_NR_NG, pM9_NR_NI,
pM9_NR_NN, pM9_NR_HD, respectively. The correct integration of the
TALEN backbone was verified by Sanger sequencing (GATC, Konstanz,
Germany). These plasmids specifically designed for producing TALENs
in P. tricornutum will be made available at Addgene (www.addgene.
org).
We created TALEN pairs with a total of 20 repeat variable di-residues
(RVD) each. Both the first RVD, which is always NG (targeting T), and
the last RVD, which is only a half monomer, are already included in
the respective plasmids. Based on these prerequisites, possible target
sites for the PtAureo1a ge ne (JGI Protein ID 49116) were generated
using the TAL Effector Nucleotide Targeter 2.0 [36] using a fixed repeat
array length of 19 per TALEN (the first NG is not counted by the soft-
ware), a spacer length of 15 to 22 bp between the two TALENs and the
P. trico rnutum Refseq ID (GCF_000150955.2) to predict possible off-
target effects. Target sites were chosen according to the following
parameters (summarized in Fig. 2): The target site should be an exon re-
gion in or upstream of the first functional domain, have no predicted off-
targets and contain few NN repeat variable di-residues (RVDs), which
lack specificity (targeting G/ A) compared to the other RVDs. Lastly,
cutting efficiency of good candidates was estimated using SAPTA
(Scoring Algorithm f or Predicting TALEN Activity; search type used:
“Score individual TALEN pair(s), optimal spacer length is assumed in
score calculation”) [37].
Cloning and insertion of the target sequences into our TALEN plas-
mids were performed as described in [33]. In short, a RVD monomer li-
brary was constructed via PCR from the plasmids pHD_v2, pNG_v2,
pNI_v2 and pNN_v2 (Addgene Kit # 1000000019, [33]). The monomers
were then digested and ligated in a Golden Gate type reaction into
hexamers, amplified via PCR a nd pu rified. Last, the three hexamers
and the plasmid containing the backbone were digested and ligated in
a second Golden Gate reaction (a summary of the TALEN assembly pro-
cedure can be found in Fi
g.
1). The resulting plasmids were sequenced
(GATC, Konstanz, Germany) to verify correct integration and order of
the 20 targeting TALE monomers.
2.2. Cultivation of algae
The P. tricornutum (Bohlin) strain UTEX646 was obtained from the
culture collection of alg ae of the University of Texas (UTEX, Austin,
USA). P. tricornutum was grown axenically in liquid F/2 medium without
added silica and 16.5‰ sal t con tent or on solid F/2 media which
contained additionally 1.2% (w/v) Bacto Agar (BD, Sparks, MD, USA).
Cells in liquid F/2 medium were cultivated in a 16 h/8 h light/dark
cycle in Erlenmeyer flasks under continuous shaking at 20 °C and an il-
lumination of 35 μmol photons m
− 2
s
− 1
(Osram Lumilux L58 W/840,
Munich, Germany). Plated cultures were cultivated under continuous il-
lumination at 75 μmol photons m
− 2
s
− 1
(Osram Biolux L30W/965).
2.3. Nuclear transformation of P. tricornutum
Nuclear transformation of P. tricornutum was performed using a Bio-
Rad Biolistic PDS-1000/He Particle Delivery System (Bio-Rad, Hercules,
CA, USA) fitted with 900/1100/1350 psi rupture disks as described pre-
viously [13,38,39].10
8
cells per plate were bombarded with 1.25 μgof
each plasmid. For selective cultivation of P. tricornutum transformants,
75 mg mL
− 1
Zeocin (Invitrogen, Carlsbad, CA, USA) and 150 mg mL
− 1
Nourseothricin (ClonNat, Werner Bioagents, Jena, Germany) were
added to the solid F/2 media [13,38].
2.4. DNA isolation
Genomic DNA was isolated using the nexttec™ 1step DNA isolation
from tissues & cells kit (Biozym, Hessisch Oldendorf, Germany) accord-
ing to the manufacturer's instructions. A cell pe llet corresponding to
10 ml of culture in mid exponential growth phase was used as starting
material. Incubation at 56 °C and 1200 rpm was done either 6 h or over-
night. Concentration of genomic DNA was measured by Nanodrop 2000
UV/VIS Spectrometer (Thermo Fisher, Schwerte, Germany).
187

2.5. Allele-specificPCR
DNA sequences for the two al leles of the PtAureo1a gene (JGI ID:
49,116) were deduced from the whole genome shotgun sequencing
(WGS) database (NCBI) by alignment of the individual sequence
reads. Alle le-specific pr imers for PCR were derived which incl ude an
allele-specific difference on the 3′ terminal bas e, thereby preventing
polymerases without proofreading func tion from amplifying the re-
spective other allele. PCR was performed using either Taq B polymerase
(Biozym, Hessisch Oldenburg, Germany) or HiDi polymerase (myPOLS,
Konstanz, Germany) according to the manufacturer's instructions. An
extension time of 4 min and an annealing temperature of 52 °C (Taq
B) or 55 °C (HiDi polymerase) were used to amplify the TALEN target
site of both PtAureo1a alleles from isolated genomic DNA using primers
Aureo1a_for and Aureo1a_rev (see Table S1). PCR products were sepa-
rated on 1% agarose gels and PCR products were isolated using the
Geneclean Turbo Kit (MP Biomedicals, Eschwege, Germany) according
to the manufacturer's instructions. Purified DNA was analyzed by Sang-
er sequencing using primer Aureo1a_for (see Table SI) (GATC, Konstanz,
Germany, or Source Bioscience, Berlin, Germany). If sequencing results
indicated mixed populations due to small insertions and/or deletions
in one or both alleles, the PCR product was sub-cloned using the
pGEM-T system (Promega, Mannheim, Germany).
2.6. Southern blotting
Isolated genomic DNA was digested using each of the following re-
striction enzymes overnight according to the manufacturer's instruc-
tions: BamHI, BsrGI, HindIII (Thermo Fisher, Schwerte, Germany).
Samples of 400 ng digested DNA were separated on 0.8% agarose gels.
The agarose gel was incubated for 10 min in denaturation solution
(0.5 M NaOH, 1 M NaCl), followed by 10 min incubation in neutraliza-
tion solution (0.5 M Tris-HCl, 3 M NaCl, pH 7.5). A dry blot was then per-
formed overnight: The gel was placed downside-up onto an acrylic glass
plate and the positively charged nylon membrane (Roche, Mannheim,
Germany; 11471240001), three layers of Whatman paper (3MM Chr,
3030917, VWR, Darmstadt, Germany) and absorbent paper were placed
on top. The blotting setup was then weighed down. The 400 bp DIG-
labeled probe was synthesized using the PCR DIG Probe Synthesis Kit
(Roche, 11636090910) using an PtAureo1a-containing plasmid [35] as
template and primers Aureo1a_probe_for/rev (see Table SI). Hybridiza-
tion occurred overnight using DIG Easy Hyb (Roche, 11603558001) at
50 °C. Post-hybridization steps were performed using the DIG Block
and Wash Buffer set (Roche, 1158 576200 1), but with a shortened
68 °C washing step (2x10min). The Anti-DIG-AP antibody (Ro che,
11093274910) was used at a 1:20.000 fold dilution; the alkaline phos-
phatase substrate used was CDP-Star (Roche, 12041677001). The blots
were developed using X-ray films (Amersham Hyperfilm ECL, GE
Healthcare , Munich, Germany) afte r 15 to 60 min incubations by a
Konica SRX-201 Developer.
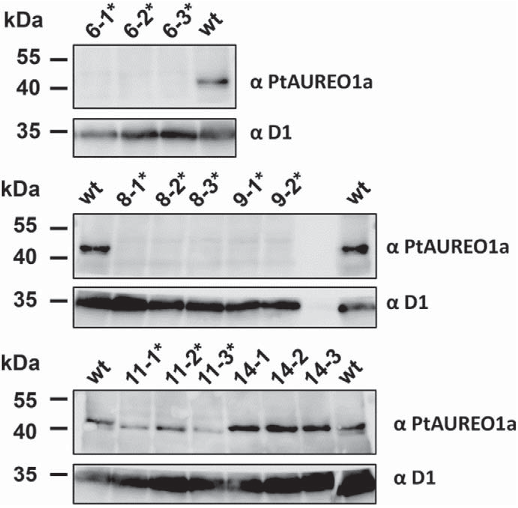
2.7. Protein isolation and immunoblotting
For protein extraction, cell pellets were resuspended in lysis buffer
(4 M urea, 1.5 M thiourea, 1% SDS, 20 m M Tris pH 8) supplemented
with protease inhibitor (Complete™ EDTA-free, Roche) according to
the manu facturer's inst ructions. A spatula tip of 1 mm, 0.5 mm and
0.1 mm diameter beads was added and the cells were lysed in a Savant
FastPrep FP120 bead mill (Thermo Scientific, Karlsruhe, Germany) six
times for 20 s with cooling on ice for 1 min between each cycle. Cell
Fig. 1. Schematic overview of the TALEN assembly process (according to [33]) and the resulting plasmids. Six monomers are assembled into hexamers corresponding to the desired target
site in a golden-gate type reaction from a library consisting of 72 monomers (18 positions of the TALEN target site and 4 different RVDs), allowing assembly of multiple fragments in the
desired order in a single step. Three of these hexamers are then ligated in a second golden gate-type reaction into the plasmid containing the first RVD (always NG) and the last half RVD
(HD, NG, NI or NN, depending on the target site). Correct insertion of the target sequence needs to be verified by colony PCR (insert size: 2.2 kbp). Clones indicated to be positive by colony
PCR should be verified by restriction digest with AfeI (expected fragment lengths: 5 kbp, 2.2 kbp and 165 bp; exemplary shown in Fig. S1) as well as sequencing of the inserted fragment.
fcpA: FcpA ( = Lhcf1) promoter; FokI: endonuclease; N-/C-term: N and C terminus, respectively; Nat: nourseothricin resistance cassette; NR: nitrate reductase promoter; RVD: repeat
variable di-residue; Sh ble: Zeocin resistance cassette.
188

debris and residual beads were removed by two centrifugation steps at
18000 g and 4 °C for 30 min.
Proteins were separated in a 10% polyacrylamide gel by SDS-PAGE
according to Laemmli et al. [40]. Each lane was loaded with protein ex-
tract of either wild type or mutant cell lines. After blotting, the nitrocel-
lulose membrane (Amersham Protran 0.1 μm NC, GE Healthcare) was
cut between 35 and 40 kDa and the top half (40–250 kDa) was used
to detect PtAUREO1a, whereas the bottom half (0– 35 kDa) was used
to detect the D1 loading control. Immunoblots using a custom-made an-
tiserum sp ecific against PtAUREO1a (Agrisera AB, Vännas, Sweden)
were performed as described in [35], whereas the D1-specific antiserum
(AS05-084, Agrisera AB) was used according to the manufacturer's in-
structions. Blots were developed using an Odyssey FC Imaging System
(Li-Cor, Bad Homburg, Germany).
2.8. Chlorophyll a determination
Chlorophyll a (Chl a) was isolated using 10% methanol and 90% ace-
tone successively and its concentration was determined spectrophoto-
metrically using the formula of Jeffrey and Humphrey [41].
2.9. Pigment extraction
Extraction of pigments and subsequent analyses via HPLC were done
as described in [42]. Samples were analyzed on a calibrated Hitachi
LaChrom Elite HPLC system equipped with a Nucleosil 120-5 C18 col-
umn (Macherey-Nagel, Düren, Germany).
2.10. Measurement of non-photochemical quenching (NPQ)
Cell suspensions in mid-exponential phase were adjusted to a Chl a
content of 1 μgml
− 1
and NPQ was measured with an AquaPen-C AP 100
(Photon Systems Instruments, Brno, Czech Republic) using light pulses
with an intensity of 2100 μmol photons m
− 2
s
− 1
applied every 20 s to
induce maximal fluorescence and 700 μmol photons m
− 2
s
− 1
of actinic
light to induce NPQ.
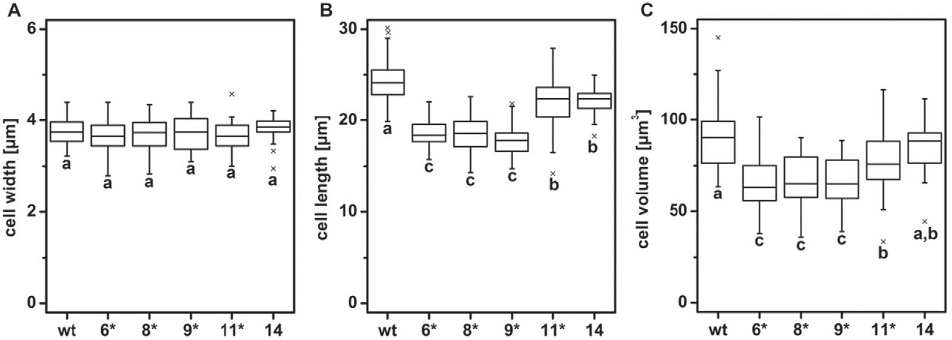
2.11. Determination of cell size by microscopy
Exponentially growing cells were analyzed using an Olympus BX51
epifluorescence microscope equipped with a Zeiss AxioCam MRm digi-
tal camera system (Carl Zeiss Microscopy GmbH, Göttingen, Germany).
Length and width of 50 cells were determined for each cell line and the
cell volume was approximated as described in [43] using the formula
V=(π/12) ∗ d
2
∗ h (d: diameter of the cell; h: length of the cell).
3. Results & discussion
3.1. Generation of the TALEN constructs
In order to perform a cost-effective and easy assembly of the TALEN
targeting sequence, we chose a system developed previously for mam-
malian systems, allowing the complete assembly and sequence verifica-
tion of individual TALEN plasmids within two weeks [33].Incontrastto
previous publications on P. tricornutum [28,30] describing the expres-
sion of TALENs in a single transformation plasmid, we decided to
clone the two TALEN backbones into two separate plasmids, one with
a constitutive fcpA promoter and sh ble gene conferring resistance to
Zeocin (pPha-T1), and one with an inducible nitrate reductase promoter
[44] and a nat gene conferring resistance to Nourseothricin (pPha-NR-
Nat). This design (sh own schematically in Fig. 1) has several advan-
tages: the expression of both TALENs from two plasmids, as compared
to a single plasmid, reduces the size of the plasmids (from 13 to 14 to
7–8 kb), which facilitates cloning procedures and, based on results
with other systems [45,46], may increase the transformation efficiency.
Furthermore, introducing two plasmids at the same time usually results
in high rates of co-transformed P. tricornutum cells even without a sec-
ond selection marker [14]. However, when using two different antibiot-
ic resistances on the two plasmids encoding the individual TALENs, a
higher selection stringency is achieved by screening for strains that
have integrated both plasmids. Larger plasmids instead are m ore
prone to random DNA double strand breaks induced by the tungsten
particles used for the particle bombardment [47]. Additionally, the in-
ducible promoter system allows switching off the expression the
TALENs once favorable mutations have been demonstrated, which de-
creases the probability of off-target DNA modifications, and allows
checking for lethal mutations by inducing the expression of the TALENs
(by exchanging ammonia by nitrate) only after the initial round of anti-
biotics selection.
The recommended workflow for designing and generating TALEN
constructs is presented in Fig. 2. In order to avoid allele-specific poly-
morphisms in the TALEN target sites, which could hinder correct bind-
ing, allele-specific gene sequences were deduced from the
P. tricornutum whole genome shotgun sequences (WGS), thereby iden-
tifying 11 different allele-specific polymorphisms in the PtAureo1a gene
for strain Pt1 (CCAP 1055/1). In the next step, a combination of two on-
line tools was used to choose the best potential target sites. TALE-NT 2.0
[36] was used to predict target sites in the gene of interest and potential
off-targets for each TALEN pair based on the P. tricornutum RefSeq se-
quence (GCF_000150955.2). A total of 191 potential TALEN pairs were
suggested within the first 700 bp of the PtAureo1a gene, of which only
29 were targeting an exon region and had no predicted potential off-
targets. These TALEN pairs were then sorted by the frequency of the
less specific NN RVD, and the in vitro cutting efficiency was estimated
by SAPTA (Scoring Algorithm for Predicting TALE(N) Activity)
[37]
.Ac-
co
rding to the SAPTA guidelines, composite scores above 30 are recom-
mended for a high rate of gene modifications. Additionally, the scores of
the individual TALENs should have similar values (e.g., 20 and 25 are
better than 5 and 40, although resulting in a similar composite score).
A SAPTA analysis of the TALEN pair chos en for PtAureo1a (left target
site: TCCCTCCTTAAGGAAGAGAA; right target site: TCGCCCAAGTGCGA
ACGAAT; spacer length: 19 bp) resulted in a composite score of 43.13
and scores for the individual TALENs of 25.44 and 20.15, respectively.
The nucleases of the TALEN pair cleave at position 679, which is part
of the leucine zipper domain of PtAUREO1a (see Fig. 3, the TALENs are
symbolized by a grey line and scissors symbols, the predicted cutting
site by a dashed line). Thus, random mutations in this area could abolish
DNA binding or may result in premature termination of protein transla-
tion. TALENs were assembled as descri bed in Materials and Methods
and verified by sequencing. We have applied the prediction tools to pre-
viously published TALEN target sites of P. tricornutum [28,30], which had
been constructed before these tools became available, and found poten-
tial off-targets for most of them. While no direct evidence is available
whether these potential off-targets are actually targeted by the TALENs
in vivo, the search tool used here is much better suited than simple
BLAST searches due to the complexity of the target sequence containing
bi-specific RVDs and a gap of variable length.
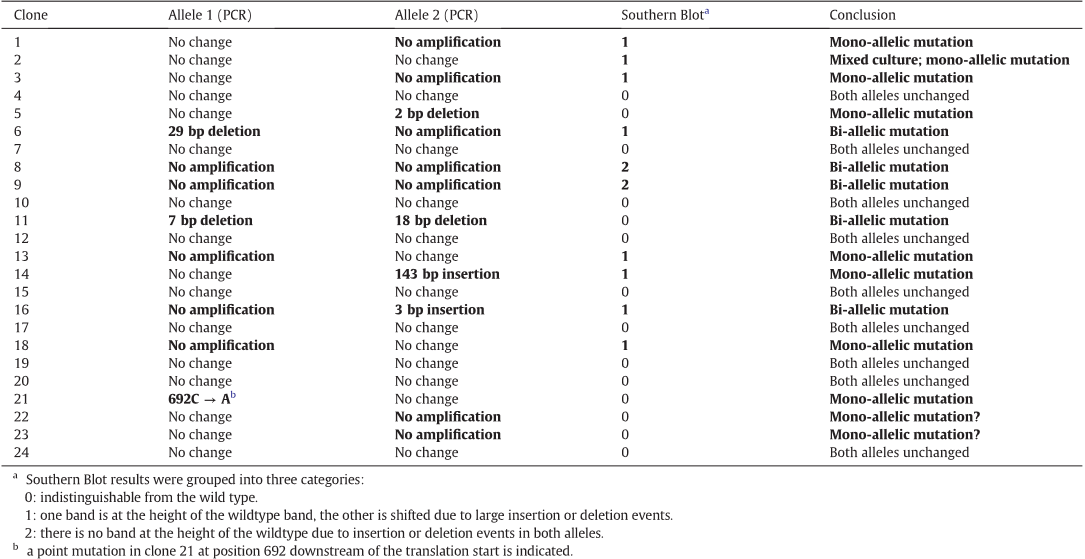
3.2. Screening of the obtained transformants and statistical evaluation
Atotalof24P. tricornutum colonies were obtained after transforma-
tion. While several allele-specific differences were identified (based on
the sequenced P. tricornutum strain Pt1 (CCAP 1055/1)), the two alleles
could not be amplified separately from Pt4 (UTEX 646), the strain used
in this study. As the availability of the genome sequence greatly facili-
tates both TALEN design and screening, the use of wild type strain Pt1
is recommended for creation of P. tricornutum knockout mutants, unless
previous work may require the use of another strain. We could solve the
problem by identifying a mixed trace peak (T/G) at position 81 of the
PtAureo1a gene in the Pt4 wild type cells as well as in the mutants,
which allowed distinguishing both alleles. Using PCR, several small de-
letions (e.g., clone 6: 29 bp deletion in allele 1) or insertions (e.g., clone
189

14: 143 bp insertion in allele 2) were detected in these transformants,
which are summarized in Table 1. No PCR products could be obtained
for cell lines 8 and 9, presumably due to large insertions or deletions
at the target site in both alleles. Furthermore, in eight transformant
cell lines only one allele could be amplified by PCR, indicating mutation
events in the respective other alleles. In case of clone 11, sub-cloning of
the PCR fragment was necessary to demonstrate small deletions in each
allele (7 and 18 bp, respectively). In summary, 10 clones were shown by
PCR and sequencing to have no targeted mutagenesis events, whereas
five clones (5, 6, 14, 16 and 21) were found to be at least mono-allelic
mutants and clone 11 was found to be a bi-allelic mutant. As not all
clones could b e reliably screened by PCR , we introduced a sec ond
screening process by employing Southern Blots of genomic DNA. The re-
striction enzymes BamHI, BsrGI and HindIII were chosen for digestion of
the genomic DNA prepared from each of the 24 mutants. For each of the
chosen restriction enzymes, Southern Blots of genomic DNA of wild
type strains showed a single band in the range of 3 to 5 kbp (see
Fig. 4). In contrast, ten of the 24 transformants (1, 2, 3, 6, 8, 9, 13, 14,
Fig. 2. Workflow and recommendations for design and assembly of TALEN constructs, as well as screening of the obtained transformants. An estimated timeframe is given for each step.
Fig. 3. Schematic drawing of PtAureo1a and its gene product PtAUREO1a including conserved domains (bZIP in red and LOV in blue). The TALEN recognition sites are indicated by a grey line
and the FokI endonuclease domains are symbolized by scissors. The predicted cutting site of the TALEN pair within the gene and its relative location within the gene product and its
conserved domains is indicated by dashed lines. The allele-specific difference of PtAureo1a at position 81 (T/G) in strain Pt4 is indicated by an arrow, an 102 bp intron region by an
inverted triangle shape and the binding site of the Southern blot probe by a magenta-colored line.
190


![Fig. 1. Schematic overview of the TALEN assembly process (according to [33]) and the resulting plasmids. Six monomers are assembled into hexamers corresponding to the desired target site in a golden-gate type reaction from a library consisting of 72 monomers (18 positions of the TALEN target site and 4 different RVDs), allowing assembly of multiple fragments in the desired order in a single step. Three of these hexamers are then ligated in a second golden gate-type reaction into the plasmid containing the first RVD (always NG) and the last half RVD (HD, NG, NI or NN, depending on the target site). Correct insertion of the target sequence needs to be verified by colony PCR (insert size: 2.2 kbp). Clones indicated to be positive by colony PCR should be verified by restriction digest with AfeI (expected fragment lengths: 5 kbp, 2.2 kbp and 165 bp; exemplary shown in Fig. S1) as well as sequencing of the inserted fragment. fcpA: FcpA ( = Lhcf1) promoter; FokI: endonuclease; N-/C-term: N and C terminus, respectively; Nat: nourseothricin resistance cassette; NR: nitrate reductase promoter; RVD: repeat variable di-residue; Sh ble: Zeocin resistance cassette.](/figures/fig-1-schematic-overview-of-the-talen-assembly-process-lkguikzl.png)