Journal of Tropical Ecology (2009) 25:415–427. Copyright © 2009 Cambridge University Press
doi:10.1017/S0266467409006075 Printed in the United Kingdom
A null-model analysis of the spatio-temporal distribution of earthworm
species assemblages in Colombian grasslands
Thibaud Deca
¨
ens
∗,1
, Juan Jos
´
eJim
´
enez† and Jean-Pierre Rossi‡
∗
Laboratoire d’Ecologie – EA 1293 ECODIV, UFR Sciences et Techniques, Universit
´
e de R ouen, F-76821 Mont Saint Aignan Cedex, France
† Instituto Pirenaico de Ecolog
´
ıa-CSIC, Avda. Regimiento Galicia, s/n. E-22700, Jaca (Huesca), Spain
‡ INRA – UMR BIOGECO, Domaine de l’Hermitage Pierroton, 69 route d’Arcachon, F-33612 Cestas, France
(Accepted 6 March 2009)
Abstract: Earthworm assemblages are usually spatio-temporally structured in mosaics of patches with different species
composition. We re-analysed results of past research carried out in Eastern Colombia to explore how interspecific
competition accounts for this pattern. In three sown pastures and three native savannas, density data matrices
were obtained from spatially explicit samplings at several successive dates, and spatio-temporal patterns of species
assemblages were described through partial triadic analyses and geostatistics. This first analysis detected assemblage
patchiness in the six plots at spatial scales ranging from 6 to 33 m. Species richness ranged from 5 to 6 species per
plot. Null models were further used to analyse niche overlap and morphometric distribution patterns at two different
scales, i.e. at the ‘plot level’ and the ‘patch level’. Seasonal and vertical niche overlaps were higher than expected by
chance at both scales, indicating high environmental constraints on assemblage membership. Within-patch overlaps
were lower than plot-scale overlaps. Biometric niche overlap was random at the plot level and was weakly lower than
that expected by chance in patches. Body weight was significantly overdispersed and constant whatever the scale,
while body length and diameter showed a similar trend within patches. These results suggest that earthworms form
distinct assemblages within patches, mainly driven by deterministic responses to competition: ecologically similar
species avoid competition through spatial segregation, whereas a minimal level of ecological segregation is required to
allow co-existence in a given patch.
Key Words: community ecology, interspecific competition, niche overlap analysis, scale dependence,
size distribution analysis
INTRODUCTION
The study of the spatial pattern of soil biota and the
factors by which they are governed is a key research
area in understanding the structure and function of soil
biodiversity and their relationships with above-ground
processes (Ettema & Wardle 2002, Ettema et al. 2000).
To date however, soil communities have been minimally
considered in spatial ecology when compared with above-
ground biota (Ettema & Wardle 2002). As an example,
despite the early recognition of the fundamental role
played by earthworms in soil processes (Darwin 1881),
the very first descriptions of their spatial distribution
were published as late as the 1950s (Boyd 1957, Guild
1951). Earthworm spatial patterns are however likely
to contribute to existing heterogeneity in soil resources
1
Corresponding author. Email: thibaud.decaens@univ-rouen.fr
and microhabitats, and to promote species co-existence
through greater resource partitioning (Lavelle 1996,
2002; Wardle 2002). In this sense earthworms could be
keystone organisms in soil faunal communities (Lavelle
et al. 2006).
In recent studies, spatial statistics have been used to
describe the horizontal spatial patterning of earthworm
assemblages at local scales (i.e. habitat surfaces of about
1 ha). In most cases, species are aggregated in patches
over ranges of 20–50 m, each of them characterized by a
dominant species assemblage that seems to be temporally
stable for about 20 mo (Deca
¨
ens & Rossi 2001, Hern
´
andez
et al. 2007, Jim
´
enez et al. 2001, 2006a; Margerie et al.
2001, Nuutinen et al. 1998, Rossi 2003, Rossi & Lavelle
1998). The determinants of these patterns are hardly
identified as they probably imply both environmental
and population or community factors operating and
interacting at different scales (Barot et al. 2007). For
instance, species-assemblage patchiness may result from

416 THIBAUD DECA
¨
ENS, JUAN JOS
´
E JIM
´
ENEZ AND JEAN-PIERRE ROSSI
species responses to the heterogeneity in plant cover and
soil properties (Margerie et al. 2001, Phillipson et al. 1976,
Poier&Richter1992), intrinsicpopulation processessuch
as reproduction rates and limited dispersal (Barot et al.
2007, Jim
´
enez et al. 2001, Rossi et al. 1997, Whalen &
Costa2003), orinterspecific competition leadingto spatial
segregation between species pairs with high niche overlap
(Jim
´
enez & Rossi 2006, Jim
´
enez et al. 2006a).
The implication of interspecific interactions, in
particular competition, in shaping the structure of natural
communities has been reported for many animals and
plants (Connell 1983, Diamond 1975, Goldberg & Barton
1992, Gotelli & McCabe 2002, Schoener 1974, Wilson
& Habiba 1995). According to the theoretical framework
of interspecific competition, two basic predictions may
be formulated (Gotelli & Ellison 2002): first, among
a set of communities or species assemblages, species
should co-occur less often than expected by chance
(EBC) (Diamond 1975, Pielou & Pielou 1968); second,
within a community or species assemblage, co-existing
species should present a lower niche overlap than EBC
(Schoener 1974). Classic examples of the latter include
limitation of similarity in body size or in multi-trait
morphology (Hutchinson 1959, MacArthur & Levins
1967, Weiher & Keddy 1995, Weiher et al. 1998). Non-
randomness in niche overlap and co-occurrence patterns
is thus considered to reflect competition constraints
on community assembly. Recently, null model analysis
has emerged as an efficient tool to identify non-
random community patterns (Gotelli 2001, Gotelli &
Graves 1996). They are pattern-generating models that
deliberately exclude a mechanism of interest (for instance
competition), and allow testing of observed data against
randomized null communities (Gotelli 2001, Gotelli &
Graves 1996). They were successfully used to highlight
and interpret, among others, non-random patterns in
body-size distribution (Feeley 2003, Gotelli & Ellison
2002) and niche overlap (Albrecht & Gotelli 2001, Hofer
et al. 2004) in different animal assemblages.
In this paper, we re-analysed with null models the
data collected in the course of different studies of the
spatio-temporal distribution of earthworm assemblages
in Colombian tropical grasslands(Deca
¨
ens 1999, Deca
¨
ens
& Rossi 2001, Jim
´
enez 1999, Jim
´
enez et al. 2006a). All
these studies described a consistent horizontal patterning
in alternated patches dominated by particular species
assemblages. We hypothesized that these patches result
from predictable assembly rules related to interspecific
competition, i.e. are a consequence of spatial exclusion
among competing species. If so, segregation should be
most evident, based on the degree of niche overlap,
at local scales (‘patch-level assemblages’, i.e. the list of
dominant species characterizing a given patch), and less
so at larger scales (‘plot-level assemblages’, i.e. the list
of species present in a given grassland plot). We thus
expected to observe two types of non-random patterns:
(1) within-patch niche overlap should be lower than
EBC and lower than plot-scale overlap (MacArthur &
Levins 1967, Weiher & Keddy 1995); (2) morphometric
distance (size ratio) between species co-existing in a given
patch should be higher and more constant than EBC, and
higher and more constant than at the plot level (Brown &
Wilson 1956, Dayan & Simberloff 2005, Gotelli & Ellison
2002, Hutchinson 1959). We tested this hypothesis
for different dimensions of the niche (seasonal activity,
vertical distribution and multi-trait morphology) and by
separately analysing different biometric traits.
An alternate hypothesis is that patchiness reflects
environmental heterogeneity and that patch-level
assemblages are composed of species sharing the adapted
traits to the patch environment. In this case, dominating
species of a given patch should present a higher niche
overlap than EBC and than at the plot-scale (Keddy 1992,
Weiher & Keddy 1995). Morphometric distance within
a given patch should also be lower and less constant
than EBC and lower and less constant than for plot-level
assemblages.
STUDY SITE
A data set was compiled from two studies carried
out at the CIAT-CORPOICA Carimagua Research
Station, in the phytogeographic unit of the well-drained
isohyperthermic savannas of eastern Colombia (4
◦
37
N,
71
◦
19
W, 175 m asl). Climate is subhumid tropical with
a mean annual rainfall and temperature of 2280 mm
and 26
◦
C, respectively (1972–1995, CIAT data).
Study plots were located in an upland area with
a well-drained silty clay Oxisol (Tropeptic Haplustox
Isohyperthermic; USDA classification), characterized by
its acidity (pH[H
2
O] = 4.5), a high Al saturation (>80%)
and low values of exchangeable cations. All the study plots
werelocated inthe same areaof theResearch Station, with
no more than 100 m between each other.
Sampling was carried out in three savanna plots and
three sown pastures. Savanna plots (Savanna 1 to 3)
were all devoid of any management, and had areas of
0.36 (Savanna 2 and 3) and 2.26 ha (Savanna 1).
Vegetation was dominated by the Poaceae Andropogon
bicornis L., Gymnopogon foliosus (Wild.) Nees, Panicum sp.,
Trachypogon sp. and Imperata brasiliensis Trin. Pasture 1
was a 1 ha and 18-y-old plot o f Urochloa decumbens
(Stapf) R.D. Webster (Poaceae) and Pueraria phaseoloides
Benth. (Fabaceae), grazed by cattle at an average stocking
rate of 1.75 Animal Unit (AU) ha
−1
(1 AU = 250 kg).
Pasture 2 was a 0.72 ha and 3-y-old plot of Urochloa
humidicola (Rendle) Morrone& Zuloaga (Poaceae), Arachis
pintoi Krap. & Greg (Fabaceae), Stylosanthes capitata Vog.
(Fabaceae) and Centrosema acutifolium Benth. ‘Vichada’

Earthworm assemblages in Colombian grasslands 417
(Fabaceae), grazed by cattle at an average stocking rate
of 2 AU ha
−1
. Pasture 3 was a 0.72 ha and 1-y-old plot
of Panicum maximum and A. pintoi, managed similarly to
pasture 2.
METHODS
Earthworm sampling
In each plot, samples were taken on a regular grid of
evenly spaced points. The dimension of the grids varied
depending on plot size: 8 × 8 sampling points each 10 m
in Pasture 1 and Savanna 1; 8 × 15 points each 5 m in
Pastures 2 and 3; 4 × 15 points each 5 m in Savanna
2 and 3. Each p lot was surveyed at different successive
dates: November 1993, 1994 and May 1995 for Savanna
1; September 1993, October 1994 and June 1995 for
Pasture 1; and every 2 mo from October 1995 to August
1997, with exception of December 1995 and July 1997,
in the other plots. Thus, the total study period for each
plot ranged from 21 to 22 mo.
At each point, a soil monolith of 40 × 40 cm (Savanna
1 and Pasture 1) or 25 × 25 cm (other plots) was
dug out down to 30-cm depth and hand sorted in the
field. Collected earthworms were identified, counted and
replaced in the monolith point with the sorted soil.
Prior to the monolith extraction, the density of the large
species Martiodrilus sp. was estimated by counting its fresh
casts at the surface of a 1-m
2
square that was centred
on the monolith (Jim
´
enez et al. 1998a). Soil monoliths
at subsequent dates were taken in points separated
about 30–50 cm from the sample of the first date. This
displacement in space was considered negligible at the
scale of the plot, and sampling coordinates were taken as
identical from one date to another.
As the characteristics of samples varied significantly
among plots, we used non-parametric regression (using
Ecosim software, Acquired Intelligence Inc. & Kesey-
Bear, http://garyentsminger.com/ecosim.htm) to verify
that differences in observed patterns were not a by-
product of different sampling procedures. This procedure
was run to test the effect of sampling grid size, distance
between sampling points, sample sizes and sampling
frequency (dependent variables) on patch spatial range
(independent variable). Non-parametric regression fits a
standard linear regression to the data set, and then uses
randomization to test the null hypothesis that the slope,
intercept or correlation coefficient equals 0. In all cases,
slope, intercept and correlation coefficient were as EBC
(r
2
= 0.00; P = 0.457 for grid size, r
2
= 0.58; P = 0.129
for distance effects, r
2
= 0.58; P = 0.140 for sample size,
r
2
= 0.58; P = 0.147 for sampling frequency). The
different sampling designs were thus assumed to address
processes at similar spatial scales.
For niche overlap calculations, we used the data sets
obtained by Jim
´
enez (1999) from a stratified random
sampling performed from April 1994 to September 1995
in the Savanna 1 and Pasture 1. In each plot, five monthly
1-m
2
monoliths were dug out down to 50 cm and hand
sorted in 10-cm increment layers. Two 20 × 20 × 20-cm
soil cores were sampled 1 m distance from the monolith;
the soil was then washed and sieved to collect small species
thatwere not efficiently collected by hand sorting (Jim
´
enez
et al. 2006b). Earthworms were fixed and stored in 4%
formaldehyde, identified and counted in the laboratory
to calculate mean population density for each sampling
month and in each soil layer. Body length (mm), weight
(g) and preclitellar diameter (mm) were measured on fixed
specimens for all specimens that were complete. Voucher
specimens of all species were deposited in the Universidad
Tecnologica de Pereira (Colombia). For both the grassland
and the savanna, we obtained three matrices describing
species according to their temporal dynamics over a
complete seasonal cycle (July 1994–June 1995), vertical
distribution and biometric traits.
Partial triadic analyses and identification of patch-level
assemblages
The partial triadic analysis (PTA) is used to analyse a
chronological series of tables that describes the same
objects with the same variables (Kroonenberg 1989, Rossi
2003, Thioulouse & Chessel 1987). It allows extraction
of the multivariate structure that is expressed through
the different dates, and describes dominant patterns
in its first axes while relegating the random noise to
further axes that are not retained for interpretation (Rossi
2003). For each plot, we used t matrices (t = number
of sampling dates), each one describing n observations
(sampling points) for p variables (species abundances).
Each PTA consisted of two successive steps: (1) The
interstructure analysis provided a global description
of the sampling points as a function of the typology
of the sampling dates. For each species and in each
plot, spatial patterns that were stable over the study
period were described by mapping the coordinates of
the sampling points on the first interstructure axis on
the sample grid. (2) The compromise analysis provided
a description of sampling points as a function of the
species typology. It was used for each plot to identify the
species assemblages that characterized similar patches at
different dates, to which we refer herein as the ‘patch-level
assemblages’. On the first compromise axis, a patch-level
assemblage was defined as a group of species displaying
coordinates of the same sign. The maps of the coordinates
of the sampling points on the first compromise axis thus
described the spatio-temporal distribution of these patch-
level assemblages.

418 THIBAUD DECA
¨
ENS, JUAN JOS
´
E JIM
´
ENEZ AND JEAN-PIERRE ROSSI
All the computations and figures were processed with
the module STATIS and other graphical modules o f the
software ADE-4 (Thioulouse et al. 1997).
Moran’s autocorrelogram
For each PTA, we tested the presence of spatial
autocorrelation in species assemblages using Moran’s
correlograms (Legendre & Fortin 1989, Sokal & Oden
1978) computed with the sample scores on the
first compromise axis (Deca
¨
ens & Rossi 2001). The
correlogram shows the changes of autocorrelation
coefficients with increasing distance. It thus indicates
the spatial range of the observed spatial patterns and
provides a test of the significance for non-randomness
(Sokal & Oden 1978). Data were allocated to nine (Pasture
1 and Savanna 1), 12 (Savannas 2 and 3) or 14 (Pasture
2 and 3) distance classes depending on the size of the
analysed matrix. Moran’s index was calculated using
the ‘Autocorr
´
elation 3.03’ module of the ‘R Package’
(Universit
´
e de Montr
´
eal, Canada), and the normality of
the data distribution was tested with a Kolmogorov–
Smirnov test before computation with the ‘VerNorm 3.0’
module of the same software. When necessary, the Box-
Cox transformation was used to reduce the asymmetry of
the frequency distribution (Sokal & Rohlf 1995).
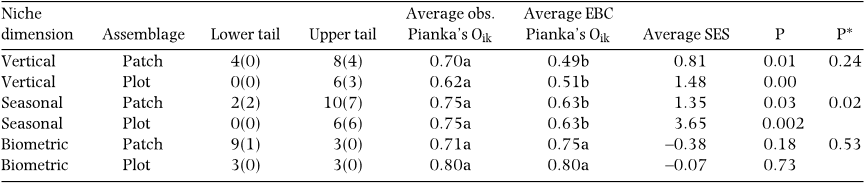
Temporal and vertical niche overlap analysis
Niche overlap analysis was undertaken for the six plot-
level assemblages and the twelve patch-level assemblages
identified by the compromise analyses. For each of them,
we built an individual matrix in which rows and columns
represented species and niche categories, respectively,
and we tested if niche overlap significantly differed from
the corresponding value under the null hypothesis (i.e.
random assemblage). We used Pianka’s index (Pianka
1973) and the Czechanowski index (Feinsinger et al.
1981). For species j and k, with resource utilizations p
ji
and p
ki
, Pianka’s overlap index of species j on species k
(O
jk
) is given by the following formula:
O
jk
= O
kj
=
n
i =1
p
ji
p
ki
n
i =1
p
2
ji
p
2
ki
For species j and k, with resource utilizations p
ji
and p
ki
,
Czechanowski overlap index (O
jk
)is:
O
jk
= O
kj
= 1.0 − 0.5 ×
n
i =1
| p
ji
− p
ki
|
The level of niche breadth was calculated with both
indices for three distinct dimensions of resource utiliza-
tion. (1) Although temporal partitioning may be relatively
uncommon in animal communities (Schoener 1974),
several examples involving invertebrates have been
described to date (Gotelli & Graves 1996). We considered
that time was of potential importance for earthworm
assemblages because the high seasonal fluctuations of
important resources, such as water and litter, may cause
diet shifts according to the time of the year. For this
analysis, we used the seasonal dynamics data. Each
individual matrix was a table where rows and columns
representedspecies andmonths respectively. Table entries
were the average number of individuals collected for each
species at a given month. (2) Niche partitioning according
to species vertical distribution is a common feature in
soil animal communities (Lavelle & Spain 2001, Wardle
2002). When foraging at different depths, earthworm
species may reduce competition by feeding on different
types of organic resources (Bouch
´
e 1977). Vertical niche
partitioning among Carimagua’s earthworms has been
suggested by Jim
´
enez & Deca
¨
ens (2000), who found that
the average living depth differed substantially among
species. To quantify and test vertical niche overlap, we
used individual matrices in which rows and columns
represented species and soil strata, respectively, and
where entries consisted of the mean number o f individuals
collected in each stratum over the total study period.
As most species were not represented below 40 cm, we
performed the analysis with the data of the first three
10-cm layers, and bulked the deeper layers into a single
‘<30 cm’ category. (3) In ecological communities,
ecologically similar species that are also morphologically
alike might not co-exist because of excessive overlap in
their resource uses (Hutchinson 1959). Consequently,
biometric traits have been widely used to quantify
the influence of competition on community assembly
(Dayan & Simberloff 2005). Biometric niche overlap
was calculated for individual matrices where rows
and columns represented species and biometric traits,
respectively, and where the entries consisted of the mean
trait values measured for the collected individuals. Here,
we used three traits (body length, weight and preclitellar
diameter) that describe earthworm external morphology
in a reliable way (Jim
´
enez 1999). To avoid any influence
of the measurement units in the index calculation, data
were previously standardized by dividing each value by
the standard deviation of the corresponding column in
the matrix.
Mean niche overlap was calculated for each patch- and
plot-level assemblage and compared with a null model
in which the observed data were randomized among
species (10 000 iterations). If competitively structured, a
given assemblage should present less niche overlap than
EBC for the dimensions of the niche that are subject to
competition. We used a randomization algorithm that
retains the niche breadth of each species, but randomizes

Earthworm assemblages in Colombian grasslands 419
which particular resource states are utilized (RA3 in
Albrecht & Gotelli 2001). It corresponds to a simple
reshuffling of each row of the matrix that assumes all
the different resource states to be equally abundant (or
usable) by all species. Calculations and tests were done
with the ‘Niche Overlap’ module of Ecosim.
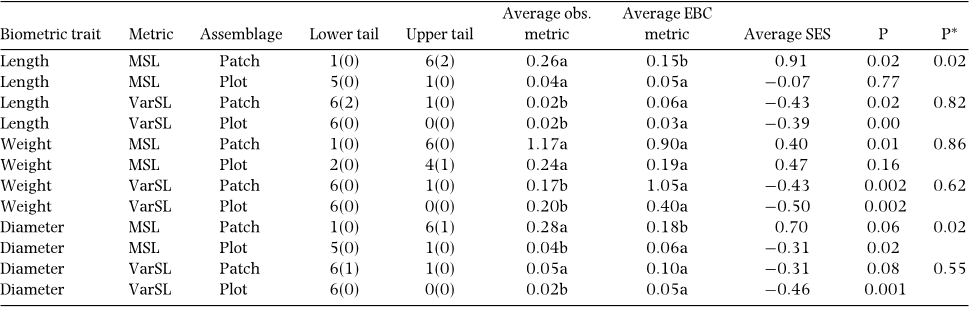
Size distribution analysis
We tested if identified assemblages presented patterns that
limit biometric similarity between co-existing species for
the three morphometric traits that were used in the niche
overlap analysis. For each trait, we calculated: (1) the
minimum segment length (MSL), which is the smallest
size difference found in all available pairs of species;
(2) the variance in segment length (VarSL) that measures
the overall tendency for the trait values to be evenly
spaced. Both metrics were successively computed after
log-transforming the data, which allowed a nalysing the
size ratio of the considered trait (Gotelli & Ellison 2002).
Observed values were calculated for all assemblages that
comprised more than two species, and were compared
with those obtained for 10 000 random assemblages. In
a competitively structured community or assemblage,
MSL and VarSL should be higher and lower than EBC,
respectively (Gotelli & Ellison 2002). We used a null model
algorithm which takes the largest and smallest species
in the assemblage to set the minimum and maximum
boundaries for the simulation. The remaining (n – 2)
species were chosen from a random, uniform distribution
within these limits. This null model assumes that in evol-
utionary time, any possible configuration of body sizes is
equiprobable within the limits imposed by the largest and
smallest species (Gotelli & Ellison 2002). Calculations and
tests were done with the ‘Size Overlap’ module of Ecosim.
Data comparisons
For each index (Pianka’s and Czechanowski O
ik
, MSL,
VarSL), we calculated the standardised effect size (SES):
SES =
(I
obs
− I
si m
)
S
si m
where I
sim
is the mean index of the simulated assem-
blages, S
sim
is the standard deviation, and I
obs
is the
observed index (Gotelli & Graves 1996). For each type of
assemblage (patch-level or plot-level) and each index, we
further calculated the average values of the observed and
simulated indices, and the average corresponding SES.
We used a permutation test to compare: (1) the average
values of the observed indices with those of simulated
assemblages; (2) the average observed values obtained in
patch-level assemblages with those obtained for plot-level
assemblages. A similar approach was used to test whether
average SES values obtained for a given set of assemblages
did differ from zero or not. Each test involved 10 000
iterations in which the data were reshuffled among the
categories to determine how much variation was expected
among the means. The null hypothesis was that the
observed variation among the means of the groups was
no greater than EBC. Calculations were performed using
the ‘Anova’ module of Ecosim.
RESULTS
Earthworm assemblage composition
A total of six species, all still undescribed and all native
from the study region, was identified in the six sampled
plots (Jim
´
enez 1999). Apart from Andiorrhinus sp., which
occurred only in Pasture 1 and Savanna 1, all species
were present in all the plots (Tables 1 and 2). Mean total
earthworm density and biomass respectively ranged from
16–25 ind. m
−2
and 1.7–4.2 g fw m
−2
in the savannas,
and 45–97 ind. m
−2
and 5.5–62.1 g fw m
−2
in the pas-
tures. Detailed studies of species assemblage composition
in the different study plots have been published previously
in Deca
¨
ens & Jim
´
enez (2002) and Jim
´
enez et al. (1998b).
The highest densities were recorded for Glossodrilus sp.
and, in the pastures, Ocnerodrilidae sp. Depending on the
Table 1. Main features of the spatio-temporal distribution of
earthworm communities in the six study plots as described by
the partial triadic analyses. CPI1 = first axis of the interstructure
analysis; CPC1 = first axis of the compromise analysis; Moran’s
P = significance level of the spatial patterns.
Plots
Number
of species
% inertia
CPI1
% inertia
CPC1
CPC1 patch
range
(metres)
CPC1
Moran’s P
Pasture 1 6 44.9 28.1 22.0 < 0.001
Pasture 2 5 20.2 34.2 22.4 < 0.001
Pasture 3 5 17.9 35.3 11.2 0.003
Savanna 1 6 38.4 33.1 33.0 < 0.001
Savanna 2 5 14.6 36.4 11.9 < 0.001
Savanna 3 5 12.9 36.6 6.0 0.002
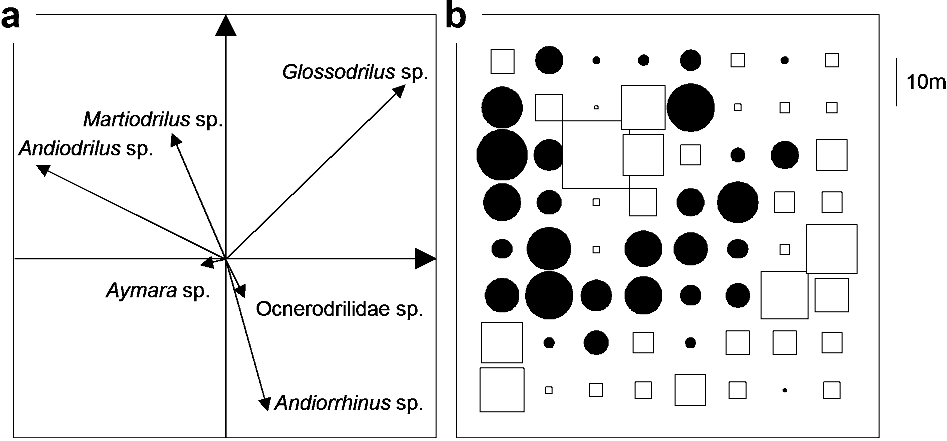
Table 2. Composition of the species assemblages in the six
study plots. For a given plot, species with the same letters
belong to the same patch-level assemblage as identified
by the first component of the compromise analysis (a =
species with positive scores; b = species with negative scores).
Species codes: And = Andiodrilus sp.; Anr = Andiorrhinus sp.;
Aym = Aymara sp.; Glo = Glossodrilus sp.; Mar = Martiodrilus
sp.; Ocn = Ocnerodrilidae.
Ocn Mar Glo And Aym Anr
Pasture 1 a b a b b a
Pasture 2 a a a b b –
Pasture 3 b b a a b –
Savanna 1 b b a b b a
Savanna 2 a b b a a –
Savanna 3 a a b b b –