




Did you find this useful? Give us your feedback










35 citations
...Admixture estimates were copied from Salzano and Sans (2014), MorenoEstrada et al. (2014) and Salazar-Flores et al. (2015)....
[...]
31 citations
29 citations
27 citations
23 citations
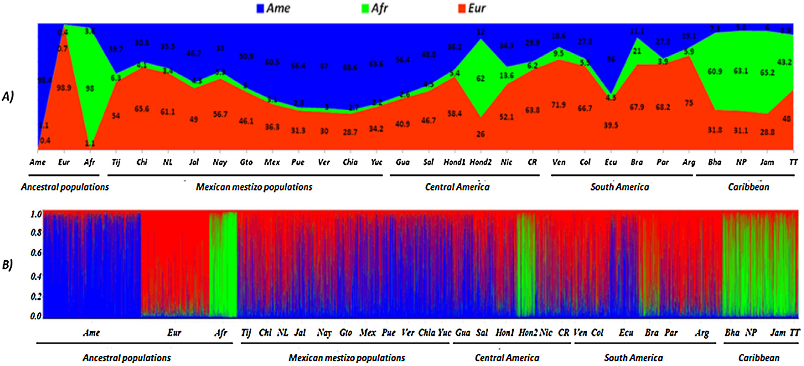
...(9), using CODIS markers, found the highest African component in Caribbean populations, and the lowest in Mexico....
[...]
542 citations
...Probably this is a result of the greater Pre-Hispanic population present in both Mesoamerica and the Andean Region in South America, as previously described (Wang et al., 2007; Rubi-Castellanos et al., 2009b)....
[...]
431 citations
...In Latin-America, hybrid populations have emerged since the European contact with the New World in 1492, and currently reflect a complex genetic structure from old and recent admixture processes (Bryc et al., 2010; Wang et al., 2008)....
[...]
...These studies have included genome-wide SNPs (Silva-Zolezzi et al., 2009; Bryc et al., 2010) and autosomal STRs in different Latin American populations (Godinho et al., 2008; Marino et al., 2006 Wang et al., 2008), and particularly CODIS-STRs in Mexican populations (Rubi-Castellanos et al., 2009a)....
[...]
...In fact, this analysis is not possible because in most of the cases we included only one population from these countries; however, these issues have been addressed in previous studies (Corach et al., 2010; de Assis Poiares et al., 2010; Marino et al., 2006; Rojas et al., 2010; Wang et al., 2008)....
[...]
...3 and 4) is closely related with the Pre-Hispanic population densities, as previously claimed (Wang et al., 2008; Rubi-Castellanos et al., 2009)....
[...]
...For anthropological purposes, their high mutation rate allows approaching historical questions such as admixture, structure, and migratory events, among others (Wang et al., 2008)....
[...]
384 citations
...(Bryc et al., 2010; Silva-Zolezzi et al., 2009)....
[...]
...These studies have included genome-wide SNPs (Silva-Zolezzi et al., 2009; Bryc et al., 2010) and autosomal STRs in different Latin American populations (Godinho et al., 2008; Marino et al., 2006 Wang et al., 2008), and particularly CODIS-STRs in Mexican populations (Rubi-Castellanos et al., 2009a)....
[...]
...In Latin-America, hybrid populations have emerged since the European contact with the New World in 1492, and currently reflect a complex genetic structure from old and recent admixture processes (Bryc et al., 2010; Wang et al., 2008)....
[...]
356 citations
...(Bryc et al., 2010; Silva-Zolezzi et al., 2009)....
[...]
...This program has been selected because results are similar to ancestry estimates based on genome-wide SNPs (Silva-Zolezzi et al., 2009), which are presumably more reliable....
[...]
...These studies have included genome-wide SNPs (Silva-Zolezzi et al., 2009; Bryc et al., 2010) and autosomal STRs in different Latin American populations (Godinho et al., 2008; Marino et al., 2006 Wang et al., 2008), and particularly CODIS-STRs in Mexican populations (Rubi-Castellanos et al., 2009a)....
[...]
...They are called Mestizos and predominantly disclose the European and Native American components, with low levels of African ancestry (<5%) (Bryc et al., 2010; Silva-Zolezzi et al., 2009)....
[...]
...The admixture component pattern obtained in Mexican Mestizo populations is similar to the previously described by genome-wide SNPs and CODIS-STRs (Rubi-Castellanos et al., 2009a; Silva-Zolezzi et al., 2009)....
[...]
312 citations
...Interestingly, two pulses of African migration to the Caribbean were described in a genomewide population study (Moreno-Estrada et al., 2013): the first pulse representing genetic component more similar to coastal West African regions involved in early stages of the trans-Atlantic slave trade; the second pulse more similar to present-day...
[...]
...Interestingly, two pulses of African migration to the Caribbean were described in a genomewide population study (Moreno-Estrada et al., 2013): the first pulse representing genetic component more similar to coastal West African regions involved in early stages of the trans-Atlantic slave trade; the…...
[...]
...Finally, the time and intensity of African slave trading (Moreno-Estrada et al., 2013) seems to be crucial to delineate the actual distribution of African ancestry in Latin American and Caribbean populations....
[...]