HOMO
-
Journal
of
Comparative
Human
Biology
66
(2015)
44–59
Contents
lists
available
at
ScienceDirect
HOMO
-
Journal
of
Comparative
Human
Biology
j
o
u
rnal
homepage:
www.elsevier.com/locate/jchb
Admixture
and
genetic
relationships
of
Mexican
Mestizos
regarding
Latin
American
and
Caribbean
populations
based
on
13
CODIS-STRs
J.
Salazar-Flores
a,
1
,
F.
Zu
˜
niga-Chiquette
b,
1
,
R.
Rubi-Castellanos
c
,
J.L.
Álvarez-Miranda
b
,
A.
Zetina-Hérnandez
b
,
V.M.
Martínez-Sevilla
a
,
F.
González-Andrade
d
,
D.
Corach
e
,
C.
Vullo
f
,
J.C.
Álvarez
g
,
J.A.
Lorente
g
,
P.
Sánchez-Diz
h
,
R.J.
Herrera
i
,
R.M.
Cerda-Flores
j
,
J.F.
Mu
˜
noz-Valle
k
,
H.
Rangel-Villalobos
a,∗
a
Instituto
de
Investigación
en
Genética
Molecular,
Centro
Universitario
de
la
Ciénega,
Universidad
de
Guadalajara
(CUCI-UdeG),
Av.
Universidad
#1115,
CP
47810
Ocotlán,
Jalisco,
Mexico
b
Laboratorio
de
Genética
Forense,
Servicios
Periciales
de
la
Procuraduría
General
de
Justicia
del
Estado
de
Baja
California,
BC,
Mexico
c
Laboratorio
de
Genética-CIR
Biomédicas,
Universidad
Autónoma
de
Yucatán
(UADY),
Mérida,
Yucatán,
Mexico
d
Escuela
de
Medicina,
Universidad
Central
del
Ecuador,
Quito,
Ecuador
e
Servicio
de
Huellas
Digitales
Genéticas,
Facultad
de
Farmacia
y
Bioquímica,
Universidad
de
Buenos
Aires,
Buenos
Aires,
Argentina
f
Equipo
Argentino
de
Antropología
Forense,
Independencia
644
–
5C,
Edif.
EME1,
Córdoba,
Argentina
g
Laboratory
of
Genetic
Identification,
Department
of
Legal
Medicine,
University
of
Granada,
Granada,
Spain
h
Genomics
Medicine
Group,
Galician
Foundation
of
Genomic
Medicine
and
University
of
Santiago
de
Compostela,
CIBERER,
Santiago
de
Compostela,
Spain
i
Department
of
Biological
Sciences,
Florida
International
University,
Miami,
FL
33199,
USA
j
Facultad
de
Enfermería,
Universidad
Autónoma
de
Nuevo
León
(UANL),
Monterrey,
Nuevo
León,
Mexico
k
Instituto
de
Investigación
en
Ciencias
Biomédicas,
Centro
Universitario
de
Ciencias
de
la
Salud,
Universidad
de
Guadalajara
(CUCS-UdeG),
Guadalajara,
Jalisco,
Mexico
a
r
t
i
c
l
e
i
n
f
o
Article
history:
Received
25
August
2013
Accepted
28
August
2014
a
b
s
t
r
a
c
t
Short
tandem
repeats
(STRs)
of
the
combined
DNA
index
sys-
tem
(CODIS)
are
probably
the
most
employed
markers
for
human
identification
purposes.
STR
databases
generated
to
interpret
DNA
profiles
are
also
helpful
for
anthropological
purposes.
In
this
work,
∗
Corresponding
author.
Tel.:
+52
392
9257112;
fax:
+52
392
9257112.
E-mail
address:
hrangel13@hotmail.com
(H.
Rangel-Villalobos).
1
These
authors
contributed
equally
to
this
work.
http://dx.doi.org/10.1016/j.jchb.2014.08.005
0018-442X/©
2014
Elsevier
GmbH.
All
rights
reserved.

J.
Salazar-Flores
et
al.
/
HOMO
-
Journal
of
Comparative
Human
Biology
66
(2015)
44–59
45
we
report
admixture,
population
structure,
and
genetic
relation-
ships
of
Mexican
Mestizos
with
respect
to
Latin
American
and
Caribbean
populations
based
on
13
CODIS-STRs.
In
addition,
new
STR
population
data
were
included
from
Tijuana,
Baja
California
(Northwest,
Mexico),
which
represents
an
interesting
case
of
elevated
genetic
flow
as
a
bordering
city
with
the
USA.
Inter-
population
analyses
included
CODIS-STR
data
from
11
Mexican
Mestizo,
12
Latin
American
and
four
Caribbean
populations,
in
addi-
tion
to
European,
Amerindian,
and
African
genetic
pools
as
ancestral
references.
We
report
allele
frequencies
and
statistical
parame-
ters
of
forensic
interest
(PD,
PE,
Het,
PIC,
typical
PI),
for
15
STRs
in
Tijuana,
Baja
California.
This
Mexican
border
city
was
peculiar
by
the
increase
of
African
ancestry,
and
by
presenting
three
STRs
in
Hardy–Weinberg
disequilibrium,
probably
explained
by
recur-
rent
gene
flow.
The
Amerindian
ancestry
in
Central
and
Southeast
of
Mexico
was
the
greatest
in
Latin
America
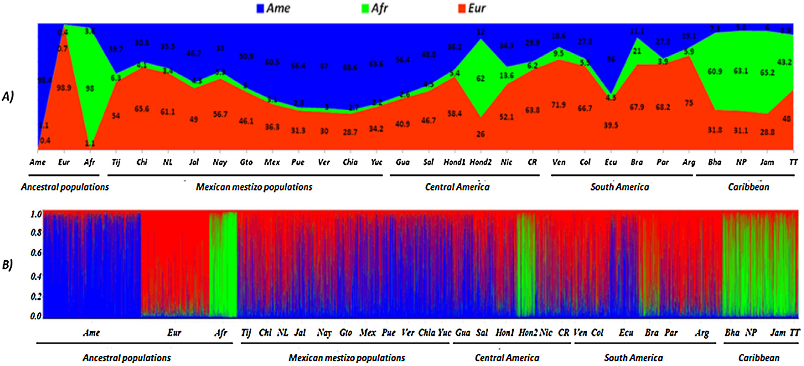
(50.9–68.6%),
only
compa-
rable
with
the
North
of
Central
America
and
Ecuador
(48.8–56.4%),
whereas
the
European
ancestry
was
prevalent
in
South
Amer-
ica
(66.7–75%).
The
African
ancestry
in
Mexico
was
the
smallest
(2.2–6.3%)
in
Latin
America
(≥2.6%),
particularly
regarding
Brazil
(21%),
Honduras
(62%),
and
the
Caribbean
(43.2–65.2%).
CODIS-STRs
allowed
detecting
significant
population
structure
in
Latin
America
based
on
greater
presence
of
European,
Amerindian,
and
African
ancestries
in
Central/South
America,
Mexican
Mestizos,
and
the
Caribbean,
respectively.
©
2014
Elsevier
GmbH.
All
rights
reserved.
Introduction
The
microsatellites
or
short
tandem
repeats
(STRs)
have
demonstrated
to
be
useful
for
linkage
and
segregation
analyses,
and
human
and
non-human
(e.g.
dogs
and
cattle)
identification.
This
is
due
to
their
elevated
heterozygosity,
genome
abundance,
high
mutation
rate,
and
simple
analysis
based
on
the
polymerase
chain
reaction
(PCR)
(Butler,
2006).
For
anthropological
purposes,
their
high
mutation
rate
allows
approaching
historical
questions
such
as
admixture,
structure,
and
migratory
events,
among
others
(Wang
et
al.,
2008).
Probably,
STRs
used
in
forensic
and
paternity
testing
are
the
most
commonly
employed
markers,
specifically
the
core
of
13
STRs
that
comprise
the
combined
DNA
index
system
(CODIS).
The
inclusion
of
CODIS-STRs
in
commercial
human
identification
kits
has
increased
the
number
of
population
databases
that
can
be
used
in
molecular
anthropology
studies
(
Butler,
2006).
The
admixture
process
presupposes
the
contact
of
ancestral
populations
that
have
been
previously
in
relative
isolation
from
each
other
and
generated
hybrid
populations,
whereas
the
population
struc-
ture
implies
differences
between
individuals
of
one
or
more
populations.
This
knowledge
is
essential
in
tasks
such
as
association
mapping,
forensic
casework,
disease
susceptibility
prediction,
wildlife
man-
agement,
and
evolutionary
studies
(Sans,
2000).
In
Latin-America,
hybrid
populations
have
emerged
since
the
European
contact
with
the
New
World
in
1492,
and
currently
reflect
a
complex
genetic
struc-
ture
from
old
and
recent
admixture
processes
(Bryc
et
al.,
2010;
Wang
et
al.,
2008).
In
Mexico,
the
European
colonization
began
in
1519,
when
Spaniards
arrived
to
the
Southeast;
they
crossed
the
cur-
rent
state
of
Tabasco
and
settled
in
Veracruz.
After
the
conquest,
about
85%
of
the
Spaniards
remained
in
conquered
territory.
Based
on
historical
records,
their
origins
are
described
as
follows:
33%
from
Andalusia,
51%
from
Leon,
Extremadura,
the
Old
and
New
Castile,
14%
from
other
regions
of
Spain,
and
some
foreigners
(6.2%)
mainly
from
Portugal
and
Genoa,
Italy
(Grunberg,
2004).
In
Mexico,
in
addition
to
the
European
component,
African
ancestry
was
incorporated
subsequently
by
means
of
slave
trad-
ing
from
various
African
countries,
such
as
Cabo
Verde,
Guinea,
and
Congo
(Aguirre-Beltrán,
1989).

46
J.
Salazar-Flores
et
al.
/
HOMO
-
Journal
of
Comparative
Human
Biology
66
(2015)
44–59
In
fact,
studies
of
mitochondrial
DNA
(mtDNA)
suggest
that
West
and
West-Central
Africa
regions
are
the
most
important
providers
of
African
ancestry
in
Central
America
and
North
America
(Salas
et
al.,
2004).
Nevertheless,
African
genes
could
have
also
arrived
in
Mexico
by
Spanish
migrants
with
Moorish
ancestry,
which
in
turn,
was
a
result
of
the
Islamic
occupation
of
Iberian
Peninsula
(Gerard
et
al.,
2006).
Currently,
most
of
the
Mexican
population
speaks
Spanish
and
is
the
result
of
admixture
between
Spaniards,
Native
Americans
and
African
populations
through
approx.
500
years.
They
are
called
Mes-
tizos
and
predominantly
disclose
the
European
and
Native
American
components,
with
low
levels
of
African
ancestry
(<5%)
(Bryc
et
al.,
2010;
Silva-Zolezzi
et
al.,
2009).
However,
a
Mexican
Mestizo
is
defined
as
a
person
born
in
the
country,
having
a
Spanish-derived
last
name,
with
family
antecedents
of
Mexican
ancestors,
at
least
back
to
the
third
generation
(Sánchez-Serrano,
1996).
A
tri-hybrid
model
has
been
used
to
explain
the
biological
diversity
of
Mexican-Mestizos,
where
specific
ancestral
com-
ponents
increase
in
different
geographical
areas:
European
in
the
North,
Amerindian
in
the
Center
and
Southeast,
and
the
African
in
the
coast.
This
model
has
been
illustrated
in
a
tripolar
diagram
where
the
edges
are
very
narrow,
indicating
a
negligible
number
of
individuals
“genetically
pure”
of
any
of
the
three
ancestries
(Gorodezky
et
al.,
2001).
The
admixture
analysis
in
Latin
America
shows
a
complex
genetic
structure
and
high
variation
of
the
Amerindian
and
European
components,
principally.
These
studies
have
included
genome-wide
SNPs
(Silva-Zolezzi
et
al.,
2009;
Bryc
et
al.,
2010)
and
autosomal
STRs
in
different
Latin
American
populations
(Godinho
et
al.,
2008;
Marino
et
al.,
2006
Wang
et
al.,
2008),
and
particularly
CODIS-STRs
in
Mexican
populations
(Rubi-Castellanos
et
al.,
2009a).
However,
during
the
last
years
further
STR
datasets
used
in
forensic
casework
have
been
reported
in
Mexican-Mestizo
populations
(Table
1),
and
the
current
inter-populational
analyses
have
not
included
the
continental
context.
In
this
study,
we
analyzed
the
ancestry,
structure,
and
genetic
relationship
of
Mexican
Mestizos
with
respect
to
Latin
America
and
the
Caribbean,
based
on
13
CODIS-STRs
population
datasets.
For
that
purpose,
we
included
reference
genetic
pools
representing
the
European,
Amerindian,
and
African
ancestries.
In
addition,
we
included
new
STR
genotype
data
of
Tijuana,
Baja
California
(BC),
the
border
city
located
in
the
North
of
Mexico
where
thousands
of
migrants
pass
to
the
United
States
of
America.
Interestingly,
this
human
mobility
comes
from
various
countries,
principally
Mexico
but
also
Salvador,
Guatemala,
Honduras,
Nicaragua,
Panama,
Costa
Rica,
and
Belize,
respectively
(Brick
et
al.,
2011).
Therefore,
Tijuana
border
city
represents
an
interesting
case
to
analyze
gene
flow
effects
in
human
populations
(INEGI,
2010).
Materials
and
methods
DNA
extraction
and
genotyping
DNA
was
extracted
from
buccal
swabs
or
peripheral
blood
by
standard
phenol-chloroform
method
from
409
unrelated
Mestizos
resident
of
the
Tijuana
City,
Mexico.
The
individuals
signed
a
written
informed
consent
according
to
the
Helsinki
Declaration.
We
amplified
15
STRs
markers
(D3S1358,
TH01,
D21S11,
D18S51,
D5S818,
D13S317,
D7S820,
D16S539,
CSF1PO,
vWA,
D8S1179,
TPOX,
FGA,
D2S1338
and
D19S433)
as
recommended
in
the
PCR
AmpFlSTR
Identifiler
kit
(Applied
Biosystems,
Foster
City,
CA).
The
amplicons
and
reference
allelic
ladders
were
analyzed
by
capillary
electrophoresis
in
the
genetic
analyzer
ABI-PRISM
310.
Results
were
interpreted
using
the
software
GeneMapper
3.2.
Statistical
analysis
In
the
population
sample
from
Tijuana,
we
estimated
the
following
forensic
parameters
with
the
software
PowerStats
(Tereba,
2001):
allele
frequencies,
heterozygosity
(Het),
power
of
discrimination
(PD),
power
of
exclusion
(PE),
polymorphic
information
content
(PIC),
typical
paternity
index
(TPI),
and
minimum
allele
frequencies
(MAF).
Furthermore,
for
each
STR
we
tested
the
Hardy–Weinberg
equi-
librium
(HWE)
and
linkage
disequilibrium
(LD)
to
check
associations
between
pairs
of
loci.
Fisher
exact
tests
based
on
3200
simulations
were
carried
out
with
the
program
Genetic
Data
Analysis
(GDA
1.1)
for
these
purposes
(Lewis
and
Zaykin,
2001).
Inter-population
analysis
was
based
on
13
CODIS-STR
datasets
including
2221
Mestizos
from
11
Mexican
populations,
1125
individuals
from

J.
Salazar-Flores
et
al.
/
HOMO
-
Journal
of
Comparative
Human
Biology
66
(2015)
44–59
47
Table
1
Geographic
region,
abbreviation,
sample
size
and
reference
of
the
admixed
population
analyzed
in
this
study.
Admixed
population
Abbr.
Sample
Reference
Mexico
(Region) Size
Tijuana
(Northwest) Tij
409
This
study
Chihuahua
(North
Center) Chi
162
Martínez-González
et
al.
(2005)
Nuevo
León
(Northeast) NL
143
Cerda-Flores
et
al.
(2002)
Jalisco
(West)
Jal
200
Rubi-Castellanos
et
al.
(2009a,b)
Nayarit
(West) Nay
200
González-Herrera
et
al.
(2010)
Guanajuato
(Center
West) Gto
200
Rangel-Villalobos
et
al.
(2010)
Mexico
City
(Center) Mex
200
Luna-Vázquez
et
al.
(2005)
Puebla
(Center)
Pue
200
Rubi-Castellanos
et
al.,
2009
Veracruz
(Center-East)
Ver
200
Rangel-Villalobos
et
al.
(2010)
Chiapas
(South) Chia
107
Sánchez
et
al.,
2005
Yucatán
(Southeast) Yuc
200
Rubi-Castellanos
et
al.
(2009a,b)
Central
America
Guatemala
Gua
200
Martinez-Espín
et
al.
(2006)
El
Salvador
Sal
200
Monterrosa
et
al.
(2006)
Honduras1
(Center-South)
Hon1
176
Matamoros
et
al.
(2008)
Honduras2
(Caribbean
coast,
Black
Garifuna) Hon2
198
Herrera-Paz
et
al.
(2008)
Nicaragua
Nic
151
Gutiérrez
et
al.
(2011)
Costa
Rica
CR
200
Rodríguez
et
al.
(2007)
South
America
Venezuela
Ven
45
Bernal
et
al.
(2006)
Colombia
Col
200
Porras
et
al.
(2008)
Ecuador
Ecu
200
González-Andrade
et
al.
(2003)
Brazil
Bra
200
Fridman
et
al.
(2008)
Paraguay
Par
181
Martínez-Espin
et
al.
(2003)
Argentina
Arg
200
Marino
et
al.
(2006)
The
Caribbean
Bahamas
Bha
162
Budowle
et
al.
(1999)
Nueva
Providencia
NP
221
Simms
et
al.
(2008)
Jamaica
Jam
160
Budowle
et
al.
(1999)
Trinidad
and
Tobago
TT
85
Budowle
et
al.
(1999)
Ancestral
Pools
Amerindian
From
Mexico
(Purepechas,
Huastecos,
Otomíes,
Tepehuas,
Amuzgos,
Chinantecos,
Choles,
Chontales,
Huaves,
Mixes,
Mixtecos,
Mazatecos,
Triquis,
Zapotecos
Zoques,
and
Mayas)
and
Ecuador
(Woaranis
and
Kichuas)
Amer
915
González-Martín
et
al.
(2008),
Quinto-Cortés
et
al.
(2010),
Ibarra-Rivera
et
al.
(2008),
González-Andrade
et
al.
(2007)
European
From
Iberian
Peninsula
(Spain
and
Portugal)
Eur
642
Camacho
et
al.
(2007),
Coudray
et
al.
(2007)
,
Lopes
et
al.
(2009)
African
From
West
Africa
(Guinea
Bissau,
Equatorial
Guinea,
and
Angola)
Afr
265
Calzada
et
al.
(2005),
Alves
et
al.
(2005)
six
Central-American
populations,
1026
persons
from
six
South-American
populations,
and
628
indi-
viduals
from
four
Caribbean
populations
(Table
1,
Fig.
1).
In
addition,
we
included
into
the
analysis
genetic
datasets
representing
the
three
main
ancestral
components
in
Latin
American
populations:
Amerindian,
European
and
African
(Table
1).
The
software
Arlequin
3.5.1.3
(Excoffier
and
Lischer,
2010)
and
the
aforementioned
population
databases
were
employed
to
perform
pairwise
comparisons,
Analysis
Molecular
of
Variance
(AMOVA),
and
F
ST
genetic
distances
were
plotted
by
multidimensional
scaling
(MDS)
with
optimum
stress
of
0.01
using
the
program
SPSS
10.0
for
Windows.
In
addition,
genetic
distances
of
Nei
(1978)
were
esti-
mated
with
the
software
GDA
1.1,
and
were
represented
in
a
neighbor
joining
(NJ)
tree
by
means
of
TreeView
3.2
(Page,
1996).
Different
population
groups
were
established
considering
genetic
and
geo-
graphical
criteria
using
the
software
SAMOVA
1.0
(Dupanloup
et
al.,
2002).
The
coordinates
(longitude
and
latitude)
were
obtained
in
Google
earth
(http://www.google.com/intl/es/earth/index.html).
The

48
J.
Salazar-Flores
et
al.
/
HOMO
-
Journal
of
Comparative
Human
Biology
66
(2015)
44–59
Fig.
1.
Geographic
location
of
the
Mexican,
Central
America,
South
American,
and
Caribbean
populations
analyzed
here.
Shadow
areas
indicate
the
Mexican
states
and
countries
included
in
this
study.
Abbreviation
meaning
can
be
consulted
in
Table
1.
components
of
admixture
were
estimated
in
individuals
and
populations
with
the
software
Struc-
ture
2.3.3
(Falush
et
al.,
2003),
with
a
burn-in-period
of
10,000
iterations
in
each
parameter
and
25
repetitions
for
each
run
(K),
using
the
mixture
model,
allele
frequency
correlation,
and
␣-value
sepa-
rated
for
populations,
with
three
populations
groups
identified
as
the
ancestral
references
(supervised
analysis).
Results
Genetic
relationships
Genetic
distances
and
pairwise
comparisons
were
estimated
between
all
Mexican-Mestizo,
Central
American,
South
American,
and
Caribbean
populations
(Suplementary
Table
1).
Nei
and
F
ST
distances
were
represented
in
a
NJ
tree
and
a
MDS
plot,
respectively
(Fig.
2A
and
B).
In
Mexico,
the
almost
per-
fect
similarity
(p
>
0.0019;
after
Bonferroni
correction)
between
populations
from
the
same
region
is
noticeable
when
they
are
separated
into
Northwest
and
Central-Southeast
regions,
and
differentiation
between
populations
from
the
opposite
region
is
visible,
supporting
the
existence
of
two
consistent
population
clusters
of
Mexican-Mestizos
(Fig.
2A).
In
Central
America,
Honduras2
(Black
Garifuna),