




Did you find this useful? Give us your feedback














1,496 citations
...Update the prior to obtain the posterior distribution (see e.g., Kruschke, 2013b, or tools on the website for Dienes, 2008: http://www.lifesci.sussex.ac.uk/home/Zoltan_Dienes/inference/bayes_normalposte rior.swf)....
[...]
...Bayes is a general all-purpose method that can be applied to any specified distribution or to a bootstrapped distribution (e.g., Jackman, 2009; Kruschke, 2010a; Lee and Wagenmakers, 2014; see Kruschke, 2013b, for a Bayesian analysis that allows heavy-tailed distributions)....
[...]
...Kruschke (2013c) recommends specifying the degree to which the Bayesian credibility interval is contained within null regions of different widths so people with different null regions can make their own decisions....
[...]
...Rules (i) and (ii) are not sensitive to stopping rule (given interval width is not much more than that of the null region; cf Kruschke, 2013b)....
[...]
...…have been ignored when they were in fact informative (e.g., believing that an apparent failure to replicate with a non-significant result is more likely to indicate noise produced by sloppy experimenters than a true null hypothesis; cf. Greenwald, 1993; Pashler and Harris, 2012; Kruschke, 2013a)....
[...]
1,190 citations
...These cases have been discussed many times in the literature, including the well-known and accessible articles by Lindley and Phillips (1976) and J....
[...]
1,172 citations
637 citations
...It should also be noted that Equation 2 is developed from a frequentist perspective, but Kruschke (2013, 2014) describes a Bayesian sample size approach based on the ROPE....
[...]
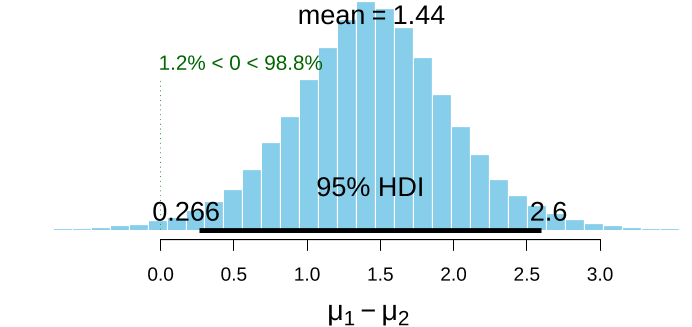
...…“actually includes the 95% of parameter values that are most credible” (Kruschke, 2013, p. 592), so “when the 95% HDI [highest density interval] falls within the ROPE, we can conclude that 95% of the credible parameter values are practically equivalent to the null value” (Kruschke, 2013, p. 592)....
[...]
...Interested readers are referred to Kruschke (2013, 2014) for such examples....
[...]
...…a Bayesian highest density interval, unlike a frequentist confidence interval, “actually includes the 95% of parameter values that are most credible” (Kruschke, 2013, p. 592), so “when the 95% HDI [highest density interval] falls within the ROPE, we can conclude that 95% of the credible parameter…...
[...]
...An attractive feature of Bayesian methods is that they often facilitate robust estimation (Kruschke, 2013)....
[...]
562 citations
272,030 citations
115,069 citations
...As a generic example, because an effect size of 0.1 is conventionally deemed to be small (Cohen, 1988), a ROPE on effect size might extend from −0.1 to +0.1....
[...]
...Importantly, the result is a different space of possible tnull values than the conventional assumption of fixed sample size, and hence a different p value and different confidence interval....
[...]
49,129 citations
...1 is conventionally deemed to be small (Cohen, 1988), a ROPE on effect size might extend from 0....
[...]
...As a generic example, because an effect size of 0.1 is conventionally deemed to be small (Cohen, 1988), a ROPE on effect size might extend from −0.1 to +0.1....
[...]
7,086 citations
6,081 citations
BEST also requires the packages rjags and coda, which should normally be installed at the same time as package BEST if you use the install.
Since BEST objects are also data frames, the authors can use the $ operator to extract the columns the authors want:> names(BESTout)[1] "mu1" "mu2" "nu" "sigma1" "sigma2"> meanDiff <- (BESTout$mu1 - BESTout$mu2) > meanDiffGTzero <- mean(meanDiff > 0) > meanDiffGTzero[1]
Once installed, the authors need to load the BEST package at the start of each R session, which will also load rjags and coda and link to JAGS:> library(BEST)The authors will use hypothetical data for reaction times for two groups (N1 = N2 = 6), Group 1 consumes a drug which may increase reaction times while Group 2 is a control group that consumes a placebo.>
If you want to know how the functions in the BEST package work, you can download the R source code from CRAN or from GitHub https://github.com/mikemeredith/BEST.Bayesian analysis with computations performed by JAGS is a powerful approach to analysis.
You can specify your own priors by providing a list: population means (µ) have separate normal priors, with mean muM and standard deviation muSD; population standard deviations (σ) have separate gamma priors, with mode sigmaMode and standard deviation sigmaSD; the normality parameter (ν) has a gamma prior with mean nuMean and standard deviation nuSD.
We’ll use the default priors for the other parameters: sigmaMode = sd(y), sigmaSD = sd(y)*5, nuMean = 30, nuSD = 30), where y = c(y1, y2).> priors <- list(muM = 6, muSD = 2)The authors run BESTmcmc and save the result in BESTout.
y1 <- c(5.77, 5.33, 4.59, 4.33, 3.66, 4.48) > y2 <- c(3.88, 3.55, 3.29, 2.59, 2.33, 3.59)Based on previous experience with these sort of trials, the authors expect reaction times to be approximately 6 secs, but they vary a lot, so we’ll set muM = 6 and muSD = 2.