Page 1/8
Complete Genome Sequence of a Novel Ourmia-like
Mycovirus Infecting the Phytopathogenic Fungus
Botryosphaeria Dothidea
Liying Sun ( sunliying@nwsuaf.edu.cn )
Northwest Agriculture and Forestry University https://orcid.org/0000-0003-2538-6096
Ziqian Lian
Northwest Agriculture and Forestry University College of Plant Protection
Subha Das
Northwest Agriculture and Forestry University College of Plant Protection
Jingxian Luo
Northwest Agriculture and Forestry University College of Plant Protection
Ida Bagus Andika
Qingdao Agricultural University School of Agriculture and Plant Protection
Research Article
Keywords: Mycovirus, Ourmia-like virus, Botourmiaviridae, Botryosphaeria dothidea, Ascomycete, Fungi,
Apple ring rot
Posted Date: May 19th, 2021
DOI: https://doi.org/10.21203/rs.3.rs-505029/v1
License: This work is licensed under a Creative Commons Attribution 4.0 International License.
Read Full License

Page 2/8
Abstract
In this study, we describe the full-length genome sequence of a novel ourmia-like mycovirus, tentatively
designated Botryosphaeria dothidea ourmia-like virus 1 (BdOLV1), isolated from the phytopathogenic
fungus,
Botryosphaeria dothidea
strain P8, associated with apple ring rot in Shanxi province, China. The
complete BdOLV1 genome is comprised of 2797 nucleotides, a positive-sense (+) single-stranded RNA
(ssRNA) with a single open reading frame (ORF). The ORF putatively encodes a 642-amino acid
polypeptide with conserved RNA-dependent RNA polymerase (RdRp) motifs, related to viruses of the
family
Botourmiaviridae
. Phylogenetic analysis based on the RdRp amino acid sequences showed that
BdOLV1 is grouped with oomycete-infecting unclassied viruses closely related to the genus
Botoulivirus
in
Botourmiaviridae
. This is the rst report of a novel (+)ssRNA virus in
B. dothidea
related to the genus
Botoulivirus
in the family
Botourmiaviridae
.
Introduction
Botryosphaeria dothidea
is a notorious canker pathogen that infects a wide range of trees worldwide [1].
This fungus is the principal causal agent of apple ring rot in China and is distributed across almost every
apple planting region [2].
B. dothidea
causes cankerous lesions on stems and brown rings on leaves and
fruits, thereby ultimately hampering the apple yield and quality [3]. While fungicides are regularly applied
for controlling this disease, the indiscriminate use of chemicals negatively impacts the environment and
poses a threat to human health [3, 4]. These concerns necessitate the development of alternative,
environmentally friendly management strategies for preventing apple ring rot.
Mycoviruses or viruses that infect fungi are present throughout all major fungal taxa [5]. Mycoviruses are
predicted to lack an extracellular phase, with their transmission occurring either vertically through conidia
or spores, or horizontally via hyphal fusion followed by cytoplasmic mixing between compatible fungal
strains [5]. Mycovirus genomes primarily consist of single- or double-stranded RNAs (ssRNA or dsRNA),
although the recent discovery of circular ssDNA mycoviruses has increased their diversity [5, 6].
In general, mycoviruses cryptically infect their hosts, although some can diminish host virulence upon
infection [7]. These viruses have the potential to be used as “virocontrol” agents for managing fungal
diseases of plants. Mycovirus-infected debilitated strains can be introduced into a eld to undergo hyphal
fusion with their virulent counterparts, making them hypovirulent upon viral transmission. The rst
successful example of such mycovirus-mediated biocontrol was using
Cryphonectria hypovirus 1
(CHV1)
to control chestnut blight caused by
Cryphonectria parasitica
[7]. Several other mycoviruses have since
been experimentally proven capable of introducing hypovirulence into their host fungi [5]. To explore
mycoviral diversity in
B. dothidea
, extensive virus hunting in this pathogen has been conducted by
numerous research groups. Such expeditions discovered several novel viruses in this fungus including
members of the families
Narnaviridae
,
Chrysoviridae
,
Fusariviridae
,
Totiviridae
,
Partitiviridae
, and
Botourmiaviridae
[8–13].

Page 3/8
Botourmiaviridae
is a recently established linear positive-sense (+) ssRNA virus family comprising four
recognized genera:
Ourmiavirus
,
Botoulivirus
,
Scleroulivirus
, and
Magoulivirus
[14]. The genus
Ourmiavirus
consists of plant-infecting viruses with encapsidated trisegmented genomes, where each
segment separately encodes a movement protein, capsid protein, and RNA-dependent RNA polymerase
(RdRp). In contrast, viruses belonging to the other three genera infect specically fungi and oomycetes
and are monosegmented with a single open reading frame (ORF) encoding an RdRp [14]
In this study, we report a novel (+) ssRNA ourmia-like mycovirus from
B. dothidea
strain 8A, which is
associated with apple ring rot in China. Sequence comparison and phylogenetic analyses suggested that
this virus is related to members of
Botoulivirus
in the family
Botourmiaviridae
and has been provisionally
named Botryosphaeria dothidea Ourmia-like virus 1 (BdOLV1).
Provenance of the virus in
B. dothidea
:
B. dothidea
strain 8A was originally isolated from an infected apple tree in Shanxi province, China. Upon
establishing pure culture, the strain was maintained on potato dextrose agar (PDA) at 25°C under dark
conditions. Strain identication was performed by internal transcribed spacer (ITS) sequencing as
described by Xu and colleagues [15]. Total dsRNA (the replicative form of the virus) was extracted from a
three-day-old mycelial culture grown on cellophane-overlaid PDA as described by Eusebio-Cope and
Suzuki and visualized by 1% agarose gel electrophoresis in 1x TAE buffer [16].
The partial cDNA sequence of BdOLV1 was initially obtained through RNA deep sequencing of ribosomal
RNA depleted total RNA from strain 8A using Illumina platform. The full-length cDNA sequence of
BdOLV1 was then obtained by amplifying its terminal regions adopting a 3' RNA ligase-mediated rapid
amplication of cDNA ends (3' RLM-RACE) method. Briey, a linker primer PC3-T7-loop (5'-p-
GGATCCCGGGAATTCGGTAATACGACTCACTATATTTTTATAGTGAGTCGTATTA-OH-3') was ligated to the 3'
ends of heat-denatured (95℃ for 4 min) viral dsRNA at 4℃ for 24 h using T4 RNA Ligase (Takara)
following the manufacturer’s instructions. The loop primer-linked puried dsRNA was then subjected to
rst-strand cDNA synthesis using SuperScript™ III Reverse Transcriptase (Invitrogen) with linker primer
PC2 (5'-CCGAATTCCCGGGATCC-3'), complementary to the 5' side of the PC3-T7-loop primer. To amplify 5'
and 3' viral terminal regions, the resulting cDNA was then amplied using 2×Es Taq MasterMix (CWBIO)
with complementary primer PC2 (5’-CCGAATTCCCGGGATCC-3’) and gene-specic primers 406R (5’-
AAACCAGGGGCGAAAGCACGAC-3’) and 2546F (CGAACTGCTGAGTCGGGGTGAT), respectively. The PCR
products were subsequently cloned using pGEM®-T Easy Vector System I (Promega). For each RACE
reaction, a minimum of three recombinant plasmids was sequenced in both directions using universal
primers M13F and M13R.
The partial viral sequence and all terminal sequences were assembled and analyzed using DNAMAN
version 9.0 (Lynnon Biosoft). The identity of BdOLV1 and its similarity to other viruses was determined
via online BLAST analyses (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The position of the ORF on the
BdOLV1 genome and its corresponding putative polypeptide were determined using the ORF nder
program (http://www.ncbi.nlm.nih.gov/gorf/org.cgi). Sequence alignments and phylogenetic analyses

Page 4/8
were performed using the MEGA version 10.1.7 software package [17]. The complete genome of BdOLV1
was submitted to GenBank under accession no. MZO73729.
Sequence properties:
The complete genome of BdOLV1 is 2797 nucleotides (nt) in length with a GC content of 54.06%
(Fig.1a). The 5'- and 3'-untranslated regions (UTRs) are 68 and 800 nt long, respectively (Fig.1a).
BdOLV1 contains a single ORF of 1929 nt, putatively encoding a 642-amino acid polypeptide with a
deduced molecular mass of 76.62 kDa (Fig.1a).
A BLASTP analysis showed that this polypeptide is related to the RdRps of several ourmia-like viruses
characterized from oomycetes and fungi. BdOLV1 RdRp was found to share 98.91%, 56.94%, and 46.51%
sequence identity with the corresponding regions of
Botryosphaeria dothidea
Ourmia-like virus (BdOLV,
unpublished partial genome sequence),
Plasmopara viticola
lesion associated ourmia-like virus 54, and
Plasmopara viticola
lesion associated ourmia-like virus 2, respectively. Despite a lack of conserved
domains in CD-Search, multiple sequence alignment of the putative RdRp region from BdOLV1 with
corresponding regions of other
Botourmiaviridae
members showed the presence of eight conserved RdRp
motifs including a highly conserved GDD signature (on motif VI) on the BdOLV1 polypeptide (Fig.1b).
Collectively, the ndings suggest that BdOLV1 is a novel ourmia-like virus in the family
Botourmiaviridae
.
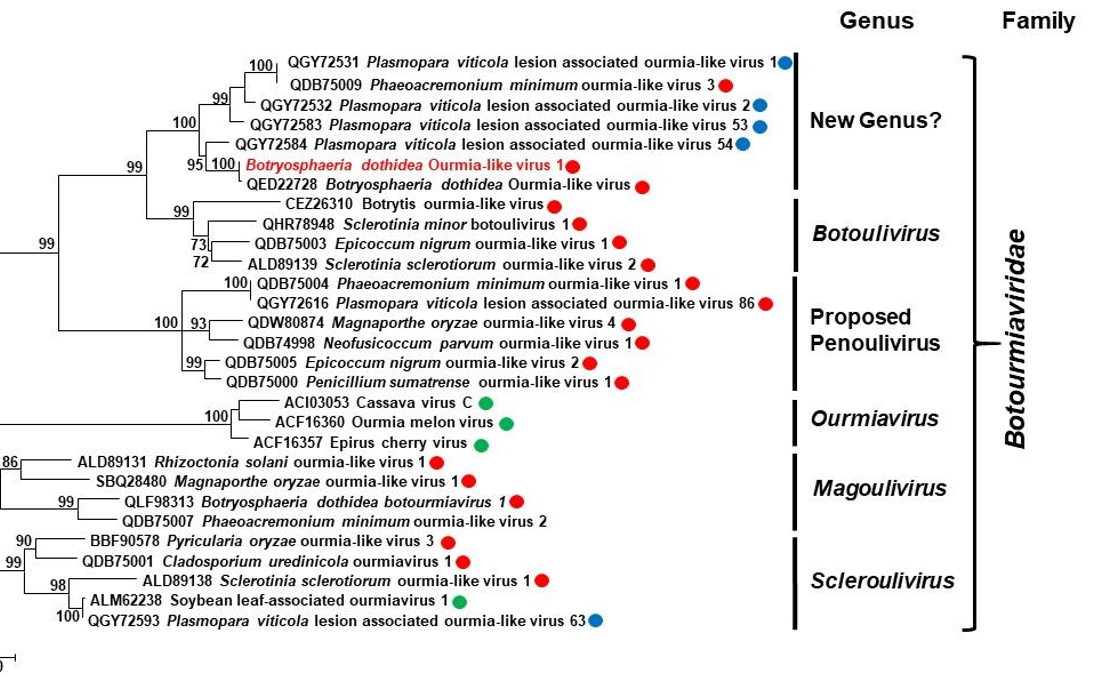
The generic identity of BdOLV1 was further determined using a Maximum Likelihood phylogenetic tree
constructed from partially conserved RdRp sequences (Fig.2). The tree topology showed that BdOLV1
grouped (100% bootstrap support) with previously reported ourmia-like viruses from the oomycete
Plasmopara viticola
and ascomycete fungi
B. dothidea
and
Phaeoacremonium minimum
(Fig.2).
Interestingly, this group of viruses showed phylogenetic relatedness to another ourmia-like viruses group
(99% bootstrap support) belonging to the genus
Botoulivirus
in the family
Botourmiaviridae
(Fig.2).
Notably, pairwise sequence alignment of full-length RdRp amino acid sequences between BdOLV1 and
Botrytis ourmia-like virus
(
Botrytis botoulivirus
), an exemplar strain of the genus
Botoulivirus
, showed
only 34.85% sequence identity, far below the current set species criteria (≤ 90%) within this genus.
Moreover, the complete RdRp sequences for members of different genera within
Botourmiaviridae
differ
by > 70% [14]. At present, it is phylogenetically dicult to conclude whether BdOLV1 and its closely
related viruses are novel species within the genus
Botoulivirus
or whether they constitute a new genus in
the family
Botourmiaviridae
.
In this study, we characterized BdOLV1 from an apple-infecting ascomycete fungus
B. dothidea
showing
no apparent disease symptoms. BdOLV1 differs from the previously characterized ourmia-like virus,
Botryosphaeria dothidea botourmiavirus 1 (BdBOV-1), which was isolated from a hypovirulent pear-
infecting
B. dothidea
strain [10]. While BdBOV-1 is phylogenetically related to the genus
Magoulivirus
,
BdOLV1 is related to the genus
Botoulivirus
in the family
Botourmiaviridae
. Interestingly, BdOLV1 shares a
close association with several ourmia-like viruses infecting oomycete
P. viticola
, suggesting a probable
exchange of such ourmia-like viruses between fungi and oomycetes. Notably, both
B. dothidea
and
P.

Page 5/8
viticola
are tree pathogens, suggesting that both organisms may have acquired such viruses from a
common source, and viruses thereafter evolved with their respective hosts.
Declarations
Acknowledgments:
We greatly thank Dr. Guangyu Sun for kindly providing research materials. This work was supported in
part by National Natural Science Foundation of China (30970163) to LY. Sun and (31970159), to IB.
Andika.
Compliance with ethical standards
Conict of interest:
All authors declare that they have no conicts of interest.
Ethical approval:
This article does not contain any studies with human participants or animals performed by any of the
authors.
References
1. Marsberg A, Kemler M, Jami F, et al (2017)
Botryosphaeria dothidea
: a latent pathogen of global
importance to woody plant health. Mol Plant Pathol 18:477–488.
2. Tang W, Ding Z, Zhou ZQ, et al (2011) Phylogenetic and pathogenic analyses show that the causal
agent of apple ring rot in China is
Botryosphaeria dothidea
. Plant Dis 96:486–496.
3. Brown-Rytlewski DE, McManus PS (2000) Virulence of
Botryosphaeria dothidea
and
Botryosphaeria
obtusa
on apple and management of stem cankers with fungicides. Plant Dis 84:1031–1037.
4. Nicolopoulou-Stamati P, Maipas S, Kotampasi C, et al (2016) Chemical Pesticides and Human
Health: The urgent need for a new concept in agriculture. Front public Heal 4:148.
5. Ghabrial SA, Castón JR, Jiang D, et al (2015) 50-plus years of fungal viruses. Virology 479–
480:356–368.
. Li P, Wang S, Zhang L, et al (2020) A tripartite ssDNA mycovirus from a plant pathogenic fungus is
infectious as cloned DNA and puried virions. Sci Adv 6:eaay9634.
7. Nuss DL (1992) Biological control of chestnut blight: an example of virus-mediated attenuation of
fungal pathogenesis. Microbiol Rev 56:561– 576
. Wang H, Liu H, Lu X, et al (2021) A novel mitovirus isolated from the phytopathogenic fungus
Botryosphaeria dothidea
. Arch Virol 166:1507–1511.