1
Ecology and Epidemiology of Wheat Curl Mite and Mite-Transmissible Viruses in 1
Colorado and Insights into the Wheat Virome 2
3
Tessa Albrecht
1
, Samantha White
1
, Marylee Layton
2
, Mark Stenglein
2
, Scott Haley
3
4
and Punya Nachappa
1
5
1
Department of Agricultural Biology, Colorado State University, 307 University Ave, Fort 6
Collins, CO 80523 7
2
Department of Microbiology, Immunology, and Pathology, College of Veterinary 8
Medicine and Biomedical Sciences, Colorado State University, Fort Collins, CO 80523, 9
U.S.A. 10
3
Department of Soil and Crop Sciences, Colorado State University, 307 University Ave, 11
Fort Collins, CO 80523 12
13
Corresponding author: Punya Nachappa; E-mail: punya.nachappa@colostate.edu 14
Keywords: wheat curl mite, wheat streak mosaic virus, triticum mosaic virus, High 15
Plains wheat mosaic virus, resistance, virome 16
Funding: Colorado Wheat Research Foundation, Colorado Wheat Administrative 17
Committee 18
19
.CC-BY-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 10, 2020. ; https://doi.org/10.1101/2020.08.10.244806doi: bioRxiv preprint

2
Abstract 20
The wheat curl mite (WCM)-transmissible wheat streak disease complex is the most serious 21
disease of wheat in the U.S. Great Plains. In the current study, we determined the genetic 22
variability in WCM and mite-transmitted viruses in Colorado and identified sources of resistance 23
in Colorado wheat germplasm to wheat streak disease complex. We identified two distinct 24
genotypes of WCM, Type 1 and Type 2 based on the ribosomal ITS1 region. Both genotypes 25
were found to co-exist throughout the wheat producing regions of Colorado. Analysis of the 26
whole genome and partial coat protein sequences revealed rich diversity of wheat streak mosaic 27
virus (WSMV) and High Plains wheat mosaic virus (HPWMoV) isolates collected from 28
Colorado, whereas triticum mosaic virus (TriMV) showed low sequence variability. Analysis of 29
WSMV isolates revealed two novel isolates and one that was 100% similar to a new variant of 30
WSMV from Kansas. Interestingly, between 2-4 genotypes of all 8 RNA segments of HPWMoV 31
were identified, which suggests new variants of emaraviruses and co-occurrence of multiple 32
strains within host populations. Several novel viruses including mycoviruses were identified for 33
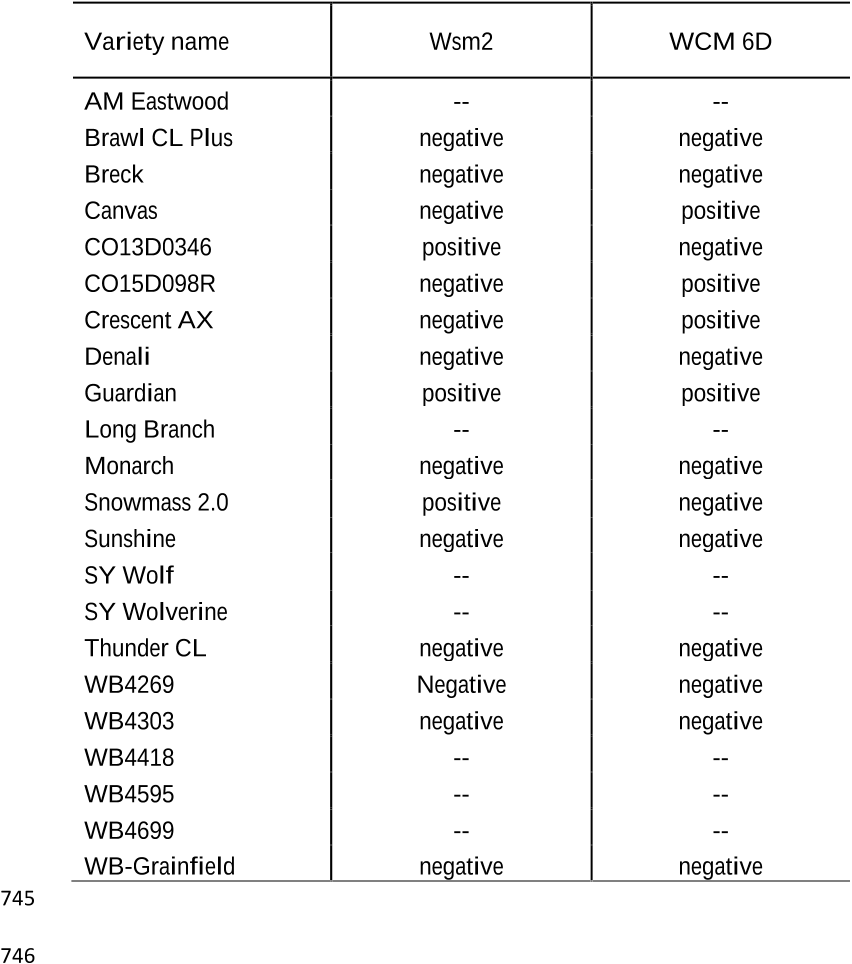
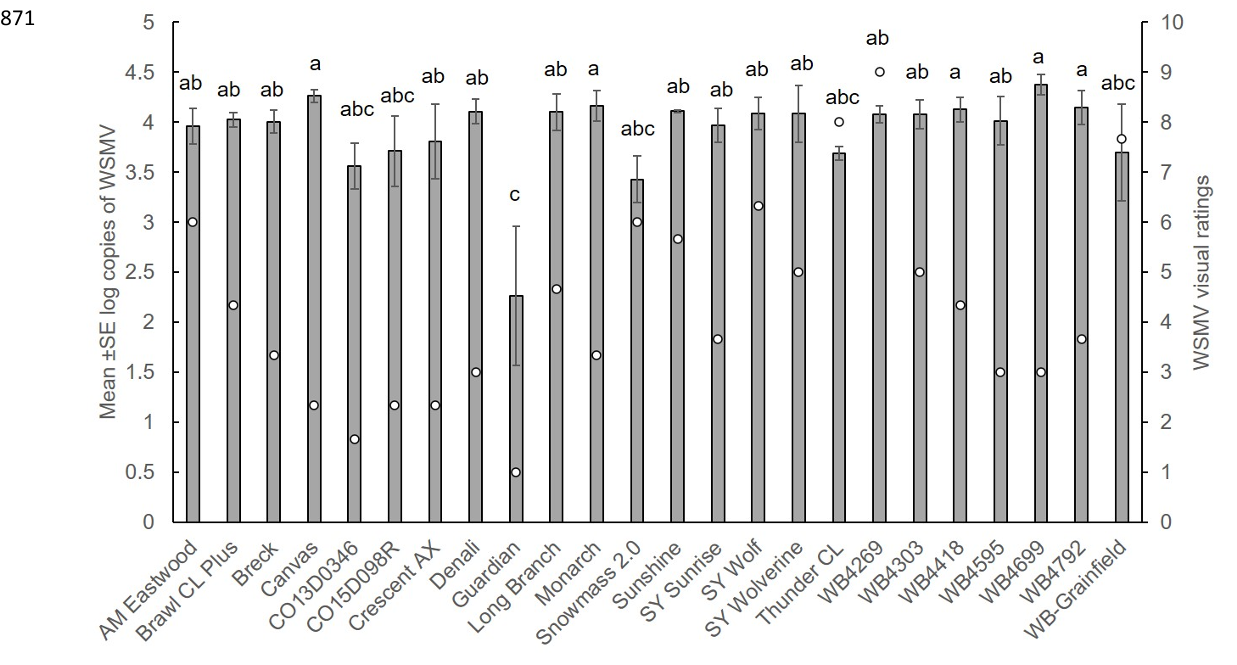
the first time in Colorado. We found variation in WSMV resistance among wheat varieties; 34
however a variety that harbored dual resistance to mite and WSMV had lower virus titer 35
compared to varieties that contained single resistance gene. This suggests that pyramiding genes 36
will ensure improved and durable resistance. Future research may be aimed at elucidating the 37
dynamics, diversity, and distribution of the new WSMV and HPWMoV isolates and their 38
responses to wheat genotypes. 39
40
Keywords: wheat curl mite, wheat streak mosaic virus, triticum mosaic virus, high plains wheat 41
mosaic virus, resistance, virome42
.CC-BY-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 10, 2020. ; https://doi.org/10.1101/2020.08.10.244806doi: bioRxiv preprint

3
Wheat (Triticum aestivum L.) is considered the most important crop in the 21
st
century as it 43
serves as a nutritional source of calories and protein in the human diet worldwide (Arzani and 44
Ashraf 2017; Curtis and Halford 2014). In the United States, wheat ranks third among field 45
crops in planted acreage, production, and gross farm receipts, behind corn and soybeans (USDA-46
ERS 2019). Among the top 10 wheat growing states, Colorado ranked 6
th
in 2019 with 2,150,000 47
acres being planted and a yield of 49 bushels per acre resulting in total production of 98,000,000 48
bushels valued at $387,100,000 (USDA-NASS 2019). The wheat curl mite (WCM), Aceria 49
tosichella Keifer (Acari: Eriophyidae) is a globally important pest affecting wheat production in 50
the Americas, Europe, and Asia (Skoracka et al. 2018). The mite causes direct damage by 51
feeding, which can reduce cereal yield (Harvey et al. 2000). But more importantly, WCM-52
transmitted viruses including wheat streak mosaic virus (family Potyviridae/genus Tritimovirus; 53
acronym WSMV) (Slykhuis 1955), triticum mosaic virus (Potyviridae/Poacevirus; TriMV) 54
(Seifers et al. 2009) and High plains wheat mosaic virus (Fimoviridae/Emaravirus; HPWMoV) 55
(Seifers et al. 1997) are among the most significant viruses in U.S. agriculture, responsible for 56
yield losses in wheat, barley, oats and rye (Burrows et al. 2009; Navia et al. 2013). Average 57
yield losses from the WCM-WSMV complex range from 5 to 7% in the US Great Plains, but 58
100% yield losses may occur in some fields (Appel et al. 2015). 59
Worldwide, the WCM has been found to be a diverse species complex with numerous 60
genetic lineages (Skoracka et al. 2018). In North America however, only two genetically distinct 61
genotypes of WCM have been characterized based on ribosomal ITS1 and mitochondrial 62
Cytochrome oxidase I/II partial sequences: Type 1, initially identified from South Dakota, 63
Kansas, Montana, Nebraska and Texas, and Type 2, from Nebraska (Hein et al. 2012). Both 64
genotypes occur in mixed populations in wheat-producing areas of the U.S. Great Plains. The 65
.CC-BY-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 10, 2020. ; https://doi.org/10.1101/2020.08.10.244806doi: bioRxiv preprint

4
two distinct genotypes demonstrate different responses to curl mite colonization (Cmc) genes; 66
Cmc1, Cmc2, Cmc3 and Cmc4 (Dhakal et al. 2017; Harvey et al. 1999) and differential viral 67
transmission efficiencies (Hein et al. 2012; McMechan et al. 2014; Seifers et al. 2002; Wosula 68
et al. 2016). For example, Type 2 is more virulent and makes wheat lines carrying the 1AL.1RS 69
(Cmc3 resistance gene) susceptible (Dhakal et al., 2017) and Type 2 mites transmit WSMV at 70
higher rates compared to Type 1 mites (Wosula et al., 2016). 71
The WSMV populations are complex as well with numerous genotypes (Robinson and 72
Murray 2013; Schubert et al. 2015), although different genotypes rarely occur in the same plant 73
(McNeil et al. 1996). In the U.S., there are two WSMV isolates, Sidney 81 and Type, sharing 74
97.6% nucleotide sequence identity, and produce similar symptoms in wheat (Choi et al. 2001; 75
Hall et al. 2001). A third isolate, El Batán, from Mexico has diverged from the American strains 76
and has 79% nucleotide sequence identity to Sidney 81 and Type (Choi et al. 2001). In contrast, 77
TriMV field populations showed minimal amounts of sequence variation suggesting that the 78
populations are very homogenous (Fuentes-Bueno et al. 2011). There is little information about 79
the phylogenetic relationships between HPWMoV isolates. There appears to be two distinct 80
groups of HPWMoV isolates within the U.S. (Stewart 2016). Currently there are three sources of 81
host resistance to WSMV - Wsm1, Wsm2, and Wsm3 (Liu et al. 2011; Lu et al. 2011; Triebe et 82
al. 1991). However, some of these resistance alleles are temperature sensitive and do not prevent 83
virus infection and replication above 18°C (Fahim et al. 2012). More recently, a novel QTL was 84
identified on wheat chromosome 6DS from the wheat cultivar, TAM112, which provides WCM 85
resistance and moderate WSMV resistance (Dhakal et al. 2018). Genes for resistance to TriMV 86
and HPWMoV have not been identified. 87
.CC-BY-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 10, 2020. ; https://doi.org/10.1101/2020.08.10.244806doi: bioRxiv preprint

5
One of the most effective ways of controlling WCM-virus complex is by planting mite 88
and disease resistant varieties; however, knowledge of mite and virus genotypes occurring in a 89
given area is critical because these genetic differences correspond to biological responses at the 90
phenotypic level (Hein et al. 2012). While Colorado is a major wheat producing state, there is no 91
information about the WCM-virus complex in the region. Moreover, little is known about 92
emerging and/or novel viruses of wheat in Colorado. Next generation sequencing is a powerful 93
tool that allows researchers to detect and characterize novel viruses (and bacterial and fungal 94
pathogens) and explore their diversity and pathogenicity in agricultural crops (Villamor et al. 95
2019). NGS is finding increased applications in revealing the viromes that contribute to the 96
disease phenotype. The term “virome” is defined as the genomes of all the viruses inhabiting a 97
specific organism or environment. In the current study, we determined the genetic variability in 98
WCM and mite-transmitted viruses in Colorado and identified sources of resistance in Colorado 99
wheat germplasm to WSMV and TriMV. In addition, we investigated the viromes of wheat from 100
four different locations in Colorado. To our knowledge, our study is among the first to report on 101
the wheat virome in the U.S. 102
103
Materials and Methods 104
Wheat Curl Mite and Plant Tissue Collection 105
Symptomatic wheat leaf tissues were collected across eastern Colorado by researchers, extension 106
agents and producers and delivered to our laboratory at Colorado State University. Plants were 107
examined using a dissecting microscope for the presence of WCMs. If present, mites were 108
transferred to healthy wheat plants of susceptible wheat varieties, Pronghorn or Hatcher at the 109
four-leaf stage or older. Plants were grown in gallon pots with 2-3 plants per pot in Promix HP
©
110
.CC-BY-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 10, 2020. ; https://doi.org/10.1101/2020.08.10.244806doi: bioRxiv preprint