1
Research Article
1
Nuclear and mitochondrial genomes of the hybrid fungal plant pathogen
2
Verticillium longisporum display a mosaic structure
3
4
Authors: Jasper R.L. Depotter
1,2†
, Fabian van Beveren
1†
, Grardy C.M. van den Berg
1
, Thomas
5
A. Wood
2‡
, Bart P.H.J. Thomma
1*‡
, Michael F. Seidl
1*‡
6
7
1
Laboratory of Phytopathology, Wageningen University, Droevendaalsesteeg 1, 6708 PB
8
Wageningen, The Netherlands
9
2
Department of Crops and Agronomy, National Institute of Agricultural Botany, Huntingdon
10
Road, CB3 0LE Cambridge, United Kingdom
11
12
† These authors contributed equally to this work
13
‡ These authors contributed equally to this work
14
*For correspondence:
15
Bart P.H.J. Thomma, Laboratory of Phytopathology, Wageningen
16
University, Droevendaalsesteeg 1, 6708 PB Wageningen, The Netherlands. Tel. 0031-317-
17
484536, e-mail: bart.thomma@wur.nl
18
Michael F. Seidl, Laboratory of Phytopathology, Wageningen
19
University, Droevendaalsesteeg 1, 6708 PB Wageningen, The Netherlands. Tel. 0031-317-
20
484536, e-mail: michael.seidl@wur.nl
21
certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprint (which was notthis version posted January 17, 2018. ; https://doi.org/10.1101/249565doi: bioRxiv preprint

2
ABSTRACT
22
Allopolyploidization, genome duplication through interspecific hybridization, is an important
23
evolutionary mechanism that can enable organisms to adapt to environmental changes or
24
stresses. This increased adaptive potential of allopolyploids can be particularly relevant for
25
plant pathogens in their quest for host immune response evasion. Allodiploidization likely
26
caused the shift in host range of the fungal pathogen plant Verticillium longisporum, as V.
27
longisporum mainly infects Brassicaceae plants in contrast to haploid Verticillium spp. In this
28
study, we investigated the allodiploid genome structure of V. longisporum and its evolution in
29
the hybridization aftermath. The nuclear genome of V. longisporum displays a mosaic
30
structure, as numerous contigs consists of sections of both parental origins. V. longisporum
31
encountered extensive genome rearrangements, whereas the contribution of gene conversion
32
is negligible. Thus, the mosaic genome structure mainly resulted from genomic
33
rearrangements between parental chromosome sets. Furthermore, a mosaic structure was also
34
found in the mitochondrial genome, demonstrating its bi-parental inheritance. In conclusion,
35
the nuclear and mitochondrial genomes of V. longisporum parents interacted dynamically in
36
the hybridization aftermath. Conceivably, novel combinations of DNA sequence of different
37
parental origin facilitated genome stability after hybridization and consecutive niche
38
adaptation of V. longisporum.
39
40
Key words: whole-genome duplication, allopolyploidy, genomic rearrangement, Verticillium
41
stem striping, Verticillium wilt, Brassica
42
certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprint (which was notthis version posted January 17, 2018. ; https://doi.org/10.1101/249565doi: bioRxiv preprint

3
INTRODUCTION
43
Whole-genome duplication (WGD) is an important evolutionary mechanism that facilitates
44
environmental adaptation (Van de Peer et al. 2017; Mallet 2005). The duplication of genomic
45
content increases genomic plasticity, leading to an augmented adaptive potential of organisms
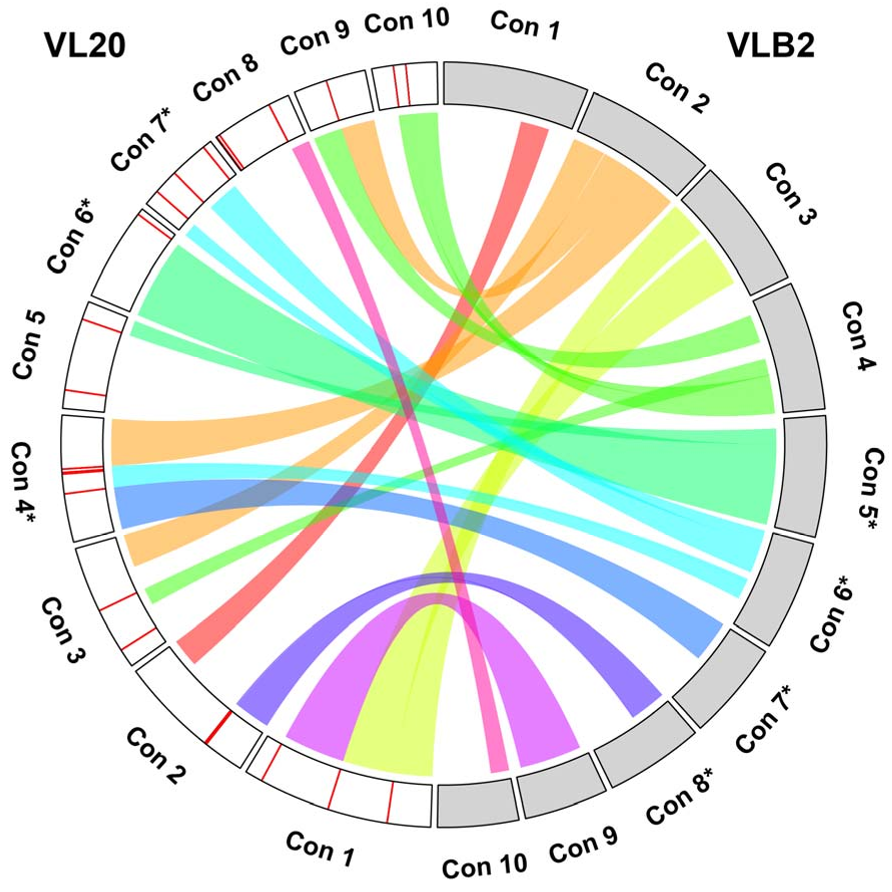
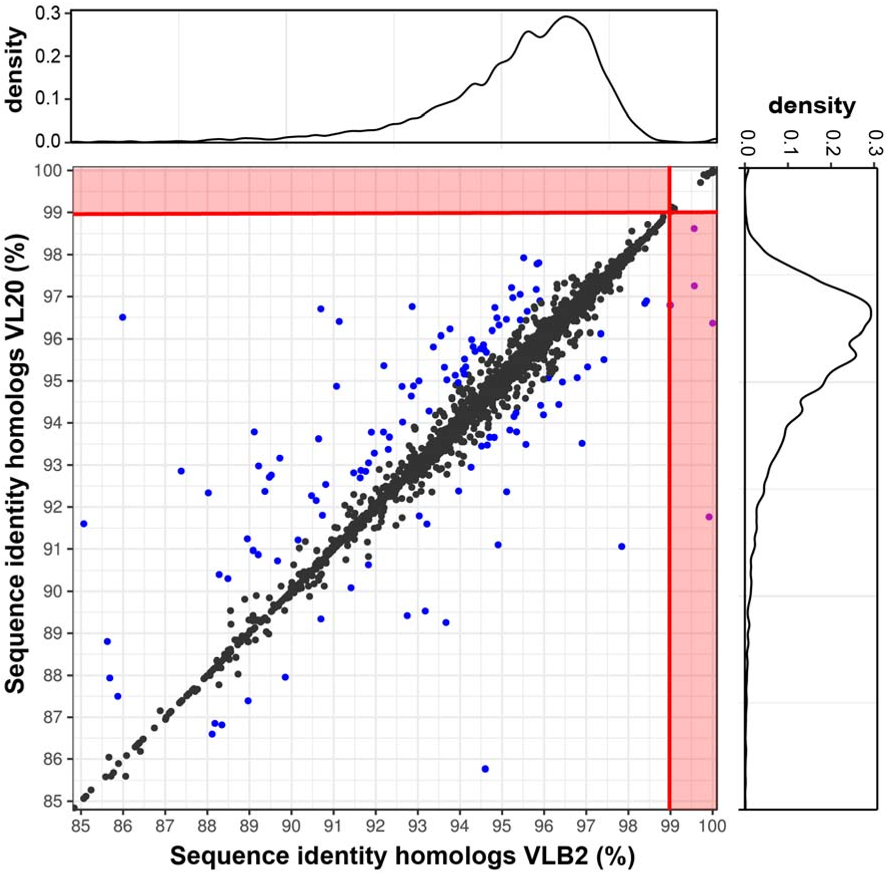
46
that underwent WGD. Consequently, polyploids have been associated with increased
47
invasiveness (te Beest et al. 2012) and resistance to environmental stresses (Lohaus & Van de
48
Peer 2016). For instance, numerous plant species that survived the Cretaceous-Palaeogene
49
mass extinction, 66 million years ago, underwent a WGD which is thought to have
50
contributed to their increased survival rates (Vanneste et al. 2014a; Vanneste et al. 2014b).
51
Both genome copies involved in WGD may have the same species origin, i.e.
52
autopolyploidization, or originate from different species as a result of interspecific
53
hybridization, i.e. allopolyploidization. In general, allopolyploids are believed to have a
54
higher adaptive potential than autopolyploids due to the increased genetic divergence between
55
the chromosome sets.
56
The impact of allopolyploidization has mainly been investigated in plants, as
57
approximately a tenth of all plant species consists of allopolyploids (Barker et al. 2015). In
58
contrast, allopolyploidization in fungi is far less intensively investigated (Campbell et al.
59
2016). Nonetheless, allopolyploidization impacted the evolution of numerous fungal species,
60
including the economically important baker’s yeast Saccharomyces cerevisiae (Marcet-
61
Houben & Gabaldón 2015). The increased adaptive potential enabled allopolyploid fungi to
62
develop desirable traits that can be exploited in industrial bioprocessing (Peris et al. 2017).
63
For instance, at least two recent hybridization events between S. cerevisiae and its close
64
relative Saccharomyces eubayanus gave rise to Saccharomyces pastorianus, a species with
65
high cold tolerance and good maltose/maltotriose utilization capabilities, which is exploited in
66
certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprint (which was notthis version posted January 17, 2018. ; https://doi.org/10.1101/249565doi: bioRxiv preprint

4
the production of lager beer that requires barley to be malted at low temperatures (Gibson &
67
Liti 2015).
68
Allopolyploid genomes experience a so-called “genome shock” upon hybridization,
69
inciting major genomic reorganizations that can manifest by genome rearrangements,
70
extensive gene loss, transposon activation, and alterations in gene expression (Doyle et al.
71
2008). These early stage alterations are primordial for hybrid survival, as divergent evolution
72
is principally associated with incompatibilities between the parental genomes (Matute et al.
73
2010). Allopolyploids benefit from a thorough re-organization where negative interactions
74
between the parental genomes are purged. Frequently, heterozygosity is lost for many regions
75
in the allopolyploid genome (Mixão & Gabaldón 2018). This can be a result of the direct loss
76
of a duplicated gene copy through deletion or gene conversion, a process where one of the
77
copies substitutes its homeologous counterpart (McGrath et al. 2014). Gene conversion and
78
the homogenization of complete chromosomes played a pivotal role in the evolution of the
79
osmotolerant yeast species Pichia sorbitophila (Louis et al. 2012). In total, two of its seven
80
chromosome pairs consist of partly heterozygous, partly homozygous sections, whereas two
81
chromosome pairs are completely homozygous. Gene conversion may eventually result in
82
chromosomes consisting of sections of both parental origins as “mosaic genomes”
83
(Stukenbrock et al. 2012). However, mosaic genomes can also arise through recombination
84
between chromosomes of the different parents, such as in the hybrid yeast
85
Zygosaccharomyces parabailii (Ortiz-Merino et al. 2017).
86
Plant pathogens are often thought to evolve while being engaged in arms races with
87
their hosts; pathogens evolve to evade host immunity while plant hosts attempt to intercept
88
pathogen ingress (Cook et al. 2015). Due to the increased adaptation potential,
89
allopolyploidization has been proposed as a potent driver in pathogen evolution (Depotter et
90
al. 2016b). Allopolyploids often have different pathogenic traits than their parental lineages,
91
certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprint (which was notthis version posted January 17, 2018. ; https://doi.org/10.1101/249565doi: bioRxiv preprint

5
such as higher virulence (Husson et al. 2015; Brasier & Kirk 2001) and shifted host ranges
92
(Inderbitzin et al. 2011b; Zeise & Tiedemann 2002). Within the fungal genus Verticillium,
93
allodiploidization resulted in the emergence of a novel pathogen on brassicaceous plants;
94
Verticillium longisporum (Inderbitzin et al. 2011b; Depotter et al. 2017b). Similar to haploid
95
Verticillium spp., V. longisporum is thought to have a predominant asexual reproduction as a
96
sexual cycle has never been described and populations are not outcrossing (Depotter et al.
97
2017b; Short et al. 2014). V. longisporum is sub-divided into three lineages, each representing
98
a separate hybridization event (Inderbitzin et al. 2011b). The economically most important
99
lineage is A1/D1 that originates from hybridization between Verticillium species A1 and D1
100
that have hitherto not been found in their haploid states. V. longisporum lineage A1/D1 is the
101
main causal agent of Verticillium stem striping on oilseed rape (Novakazi et al. 2015) and its
102
economic importance as emerging pathogen is increasing worldwide (Depotter et al. 2017a).
103
A recent study revealed that lineage A1/D1 can be further divided into two genetically distinct
104
populations, which have been named ‘A1/D1 West’ and ‘A1/D1 East’ after their geographic
105
occurrence in Europe (Depotter et al. 2017b). Nevertheless, both populations were shown to
106
originate the same hybridization event (Depotter et al. 2017b).
107
V. longisporum is assumed to have largely conserved its allodiploid state as the sizes
108
of its sub-genomes resemble those of haploid Verticillium spp. (Depotter et al. 2017b; Shi-
109
Kunne et al. forthcoming; Fogelqvist et al. 2018). Nevertheless, not all genes are present in
110
heterozygous copies, as its nuclear ribosomal internal transcribed spacer region is derived
111
only from one of the parents (Inderbitzin et al. 2011b). Here, we investigated the evolution of
112
the allodiploid genome of V. longisporum and determined to what extent heterozygosity is
113
lost.
114
certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission.
The copyright holder for this preprint (which was notthis version posted January 17, 2018. ; https://doi.org/10.1101/249565doi: bioRxiv preprint