




Did you find this useful? Give us your feedback
![Figure 1: Definition of functional and effective connectivity. (A) Classification of causal (directional link), indirect (multi-neurons pathway) and apparent (functional coupling due to common input) connectivity. (B) Classification of most common connectivity inference methods in terms of causality and detection of direct links. On the x -axes, the graph shows a scale of causality which refers to the ability of a given connectivity method to infer or not the directionality of the functional connections between neurons. On the y-axes, the graph visually quantifies the capabilities of one approach to detect direct links between neurons by identifying and discarding multi-neurons connections and apparent ones. Indicators that are acausal and do not infer direct links can only report about functional connectivity (light yellow); indicators which contain information about direction of interaction and direct neuron-to-neuron communication are close to the inference of effective connectivity (orange color). Most common model-free techniques are indicated with black dots. Model-based methods are reported with blue triangles and their ability to infer effective connectivity is negatively weighted by the impossibility to test their performances. The super-selective correlation approach we propose is reported in black like the other model-free methods; a diamond signal is used to emphasize the fact that, although being model-free, it aims at inferring effective connections. The graph visually summarizes results of comparisons between connectivity methods from review papers [14, 76, 19] and do not contain precise quantitative information about the differences.](/figures/figure-1-definition-of-functional-and-effective-connectivity-2yx7qrh7.png)








1,472 citations
473 citations
176 citations
[...]
2 citations
872 citations
851 citations
...Finally, we studied the integration and segregation properties of the networks [60] by computing the average Mean Path Length and Mean Cluster Coefficient (CCo)....
[...]
834 citations
...Among them, dynamic causal modeling (DCM) [29] and structural equation modeling [30] variants have shown best performances (Figure 1)....
[...]
752 citations
...In-vitro cultures of primary neurons can self-organize into networks that generate spontaneous patterns of activity [1, 2, 3], in some cases resembling aspects of developing brain circuits [4, 5]....
[...]
736 citations
...Methods such as correlation [34], coherence [35, 36, 37], mutual information [31, 38, 39], phase and generalized synchronization [39, 40], and joint entropy [31] describe only statistical dependencies between recorded neurons without carrying any information of causality or discriminating direct and indirect effects....
[...]
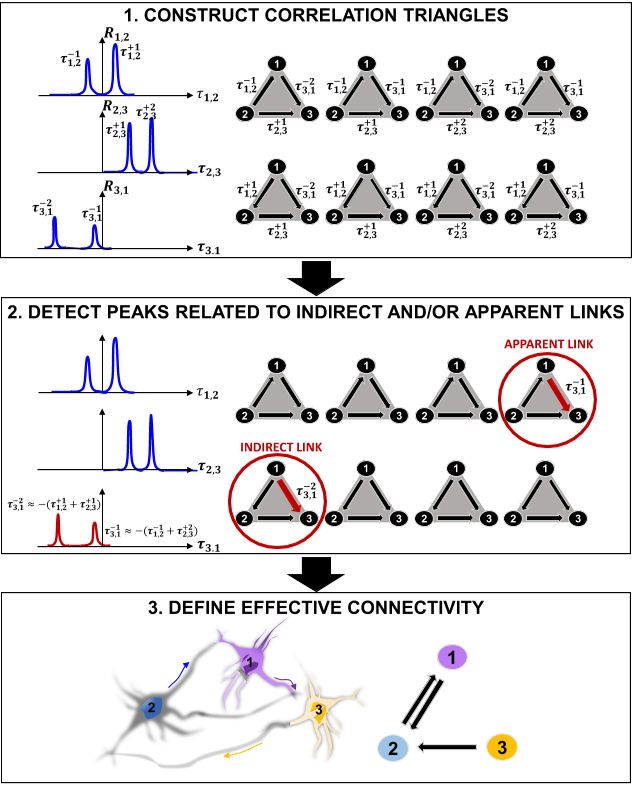
In addition, it has good scaling capabilities and can be further generalized to any kind of network, thus allowing to target different problems in intact neurons, synthetic models as well as in vitro and in vivo systems. Furthermore, it will be broadly applicable to experimental techniques for neural activation and recording, increasing its utility for the analyses of spontaneous neural activity patterns, as well as neuronal responses to pharmacological perturbations and electrical and optogenetic stimulations [ 70, 26, 38, 41, 3 ]. As novel electrophysiology technologies come online and are validated, the methods the authors presented here will be in an immediate position to take advantage of them, resulting in fundamental improvements in spatial resolution and reconstruction accuracy. Furthermore, their algorithm can be further extended, improved, and possibly integrated with already in-use techniques to overcome important limitations such as the detection of inhibitory connections and the inference of effective connectivity in the bursting regime.