




read more










BEST also requires the packages rjags and coda, which should normally be installed at the same time as package BEST if you use the install.
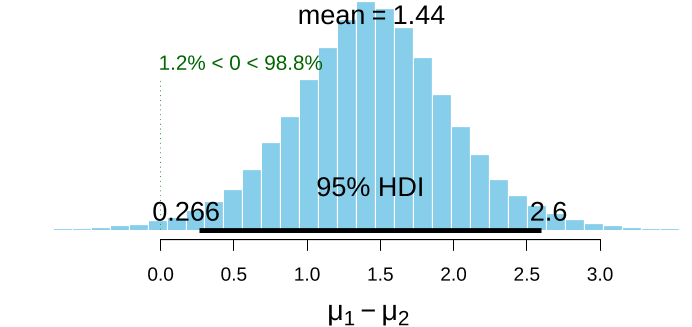
Since BEST objects are also data frames, the authors can use the $ operator to extract the columns the authors want:> names(BESTout)[1] "mu1" "mu2" "nu" "sigma1" "sigma2"> meanDiff <- (BESTout$mu1 - BESTout$mu2) > meanDiffGTzero <- mean(meanDiff > 0) > meanDiffGTzero[1]
Once installed, the authors need to load the BEST package at the start of each R session, which will also load rjags and coda and link to JAGS:> library(BEST)The authors will use hypothetical data for reaction times for two groups (N1 = N2 = 6), Group 1 consumes a drug which may increase reaction times while Group 2 is a control group that consumes a placebo.>
If you want to know how the functions in the BEST package work, you can download the R source code from CRAN or from GitHub https://github.com/mikemeredith/BEST.Bayesian analysis with computations performed by JAGS is a powerful approach to analysis.
You can specify your own priors by providing a list: population means (µ) have separate normal priors, with mean muM and standard deviation muSD; population standard deviations (σ) have separate gamma priors, with mode sigmaMode and standard deviation sigmaSD; the normality parameter (ν) has a gamma prior with mean nuMean and standard deviation nuSD.
We’ll use the default priors for the other parameters: sigmaMode = sd(y), sigmaSD = sd(y)*5, nuMean = 30, nuSD = 30), where y = c(y1, y2).> priors <- list(muM = 6, muSD = 2)The authors run BESTmcmc and save the result in BESTout.
y1 <- c(5.77, 5.33, 4.59, 4.33, 3.66, 4.48) > y2 <- c(3.88, 3.55, 3.29, 2.59, 2.33, 3.59)Based on previous experience with these sort of trials, the authors expect reaction times to be approximately 6 secs, but they vary a lot, so we’ll set muM = 6 and muSD = 2.