
1
Use of a Fluorinated Probe to Quantitatively Monitor Amino Acid
Binding Preferences of Ruthenium(II) Arene Complexes
George S. Biggs,
†
Michael J. O’Neill,
†, ‡
Pablo Carames Mendez, Thomas G. Scrase, Yulu
Lin, Amzar Muzani Bin-Maarof, Andrew D. Bond, Sally R. Boss* and Paul D. Barker*
University of Cambridge, Chemistry Department, Lensfield Road, Cambridge CB2 1EW,
UK
‡
Faculty of Science and Engineering School of Mathematics and Physical Sciences,
University of Hull, Hull, UK
†
These authors contributed equally to this project.
ABSTRACT:
In order to address outstanding questions about ruthenium complexes in complex
biological solutions,
19
F NMR spectroscopy was used to follow the binding preferences
between fluorinated Ru
II
(h
6
-arene)(bipyridine) complexes and protected amino acids and
glutathione. Reporting what ruthenium compounds bind to in complex environments has
so far been restricted to relatively qualitative methods, such as mass spectrometry and X-
ray spectroscopic methods; however, quantitative information on the species present in
the solution phase cannot be inferred from these techniques. Furthermore, using
1
H NMR,
in water, to distinguish and monitor a number of different complex Ru
II
(h
6
-arene) adducts
forming is challenging. Incorporating an NMR active heteroatom into ruthenium
organometallic complexes provides a quantitative, diagnostic ‘fingerprint’ to track solution-
phase behaviour and allow for unambiguous assignment of any given adduct. The
resulting
19
F NMR spectra show for the first time the varied, dynamic behaviour of
organoruthenium compounds when exposed to simple biomolecules in complex mixtures.
The rates of formation of the different observed species are dramatically influenced by the
electronic properties at the metal, even in a closely related series of complexes in which
only the electron-donating properties of the arene ligand are altered. Preference for
cysteine binding is absolute: the first quantitative solution-phase evidence of such
behaviour.
INTRODUCTION:
One motivation for using ruthenium in medicinal compounds is to introduce a drug-target
interaction for which the bonding energy is between that of true non-covalent
intermolecular interactions and a covalent bond.
1–3
In principle, tuning the strength of a
metal-target coordination bond could be used to control the dissociation rate of a drug-
target interaction in a way that is orthogonal to that provided by conventional medicinal
chemistry. Consequently, a raft of studies have begun to explore the binding preferences
of simple ruthenium organometallic complexes for biomolecular targets, with many of
these compounds demonstrating promising anticancer activity.
4–6
The ruthenium complexes that have entered clinical trials, NAMI-A and KP1019 have
received significant attention with respect to their distributions within plasma and in cells.
They are characterized by having a number of ligands we would class as exchangeable in
aqueous solution on a timescale relevant to cellular processes. These complexes are very
promiscuous in complex biological mixtures, with little understanding of their modes of
action or final cellular speciation.
7–9

2
Ruthenium(II) arene complexes incorporating a wide range of different ligands have also
been studied extensively in this context.
10–16
In comparison with Ru
III
complexes, the
Ru
II
(h
6
-arene) bioactive scaffold has led to increased control over the biomolecular targets
of ruthenium complexes. RAPTA-type complexes, [Ru(arene)(PTA)X
2
] preferentially bind
to proteins
17,18
whereas RAED-type complexes [Ru(arene)(en)Cl] (en = 1,2-
ethylenediamine) preferentially bind to DNA; this binding can be enhanced through
extended p-systems.
19,20
Although these complexes have shown promising anticancer
activity, they are still highly promiscuous and there is little insight into their complete
cellular speciation.
The dynamic nature of the cellular concentrations and accessibility of biomolecules
combined with the characteristically slow ligand exchange rates associated with Ru(II)
arene complexes,
21
makes understanding the relationship between speciation of the metal
complexes and cellular response problematic. A direct read out of what the ruthenium
compounds are bound to in biological environments remains challenging and has so far
been restricted to relatively qualitative methods, such as mass spectrometry
22–24
and X-ray
spectroscopic methods.
25–27
NMR spectroscopy is a quantitative, sensitive, direct reporter for solution behaviour.
However, using 1-Dimensional
1
H NMR to follow the speciation of ruthenium complexes
has proved testing for anything other than the simplest examples. The large number of
observable proton signals together with their chemical similarity and hence proximity in the
spectra severely restricts the usefulness of
1
H NMR in this context. 2-Dimensional NMR
experiments can overcome this complexity; however, these experiments are often time
consuming, with high sample concentration necessary, if temporal resolution is required.
31
P NMR has been used extensively for following the metabolic state of phosphate esters
in vivo.
28–30
15
N and
13
C are commonly used to examine the structure and dynamics of
proteins in vivo.
31
19
F NMR can also be used as a sensitive probe of specific metabolic signals when a
fluorine atom can be incorporated into a small or large biomolecule as a reporter.
32,33
The
use of ligand-based fluorine NMR screening methods to rapidly identify biologically active
fluorinated organic compounds has demonstrated the usefulness of being able to detect a
19
F NMR signal in complex environments.
34,35
Furthermore, incorporating fluorine atoms into lead drug compounds, has proven to be a
popular way to improve the pharmacological properties of organic-based drugs, with
approximately 20% of all currently available drugs on the market containing at least one
fluorine atom.
34
Recently, a fluoride ligand has been incorporated into the axial position of
a Pt(IV) anticancer pro-drug to enhance stability and cytotoxicity.
36
Therefore, we believe
that incorporating fluorine substituents into Ru
II
arene complexes can be greatly beneficial
in probing their reactivity in complex mixtures, as well as a means of altering activity.
Throughout this study, we have used
19
F NMR to gather quantitative information, such as
kinetic parameters and percentage formation of the species present in solution, with mass
spectrometry used to support the assignment of species. This combined approach can
greatly simplify observing the binding preferences of Ru(II) arene in complex biological
mixtures. The purpose here, was not to develop a new bioactive ruthenium organometallic
compounds, but to explore a quantitative spectroscopic method for quickly and accurately
assigning the speciation of ruthenium organometallics in complex solutions.

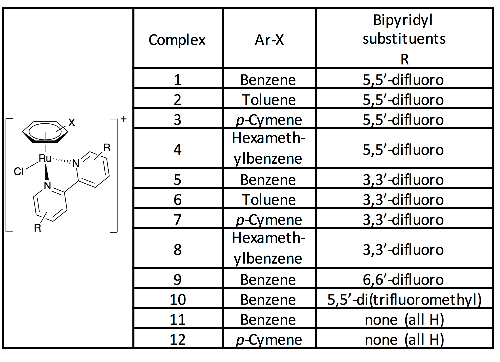
3
Table 1: Complexes synthesized and studied in this work, isolated and used as the
hexafluorophosphate salts.
Incorporating the fluorine atoms into a bipyridyl ligand, which is stable to dissociation,
enabled at least two fluorines in chemically similar environments to be introduced to each
complex. We synthesised four fluorinated 2,2’-bipyridyl derivatives; 3,3’, 5,5’, 6,6’-difluoro
and 5,5’-trifluoromethyl. To counteract the electron withdrawing effect of the fluorinated
ligands on the metal, we also varied the arene ligand, including more electron-donating
substituted arenes by way of compensation. The full set of complexes reported is listed in
Table 1, and Figure S1.
Some of these complexes provide excellent sensitivity to the metal coordination
environment and dispersion of signals that have allowed us to resolve the surprising
number of different species present. The sensitivity we report suggests that this approach
could be developed for use in vivo and we have made attempts to test that. We also show
that the rates of formation of different species are dramatically influenced by the electronic
properties at the metal, even in a closely related series of complexes in which only the
electron-donating properties of the arene ligand are altered.
RESULTS AND DISCUSSION
A number of literature preparations are available for the preparation of functionalised
bipyridines,
37,38
however, after achieving relatively poor yields using copper catalysed,
Ullman-type coupling reactions, the fluorinated bipyridines were synthesised using an
adapted palladium-catalysed homocoupling procedure in poly(ethylene) glycol (see
Experimental Section).
39
We found that all except the 4,4’-difluorobipyridyl derivative could
be made reliably in good yield; we have not pursued the 4,4’ derivative any further. The
synthesis of the ruthenium complexes [1] – [12] was achieved by reacting the appropriate
ruthenium arene dimer with a stoichiometric amount of the chosen bipyridyl derivative.
These complexes were characterised by NMR, ESI-MS, elemental analysis and X-ray
diffraction.
Single crystals suitable for X-ray diffraction for complexes [1] – [10] were grown via slow
diffusion of diethyl ether into a saturated solution of the complex in acetone. The X-ray
crystallographic data for all complexes are given in Tables S2-4. The novel complexes [1]-
[10] adopt a pseudo-octahedral piano stool configuration. A representative analysis of key
bond lengths and angles for the 5,5’-fluorinated bipyridine series is summarised in Table
S1. Complexes [1] and [2] have shorter Ru-Cl bond lengths than complexes [3] and [4]

4
which could provide a hint towards the lability of the chloride when other nucleophiles are
present, but these small differences cannot be used to inform solution based behavior.
Speciation in Aqueous Solutions
Before exposing the ruthenium complexes to biomolecules, they were monitored in
deuterated phosphate buffer, pD = 7.2 and 1% DMF which leads to an equilibrium
between chloro, aquo and phosphate adducts, Figure 1a. In order to be able to see as
many relevant species as possible, high concentrations of the complexes under study
were needed so a small amount of DMF was used, to enhance solubility. We have seen
no evidence of DMF coordinating under these conditions. For the fluorinated complexes
[1] - [8] and [10], three singlet peaks are observed in the
19
F NMR spectra, as represented
in Figure 1b. The behaviour of complex [9], the 6,6’-bipyridyl derivative, was significantly
different suggesting that the proximity of the fluorines to the metal impacted upon the
ligand exchange properties. For [11]-[12],
1
H NMR spectra were used to resolve the
speciation through the diagnostic metal-coordinated arene signals.
40
Figure 1: Figure 1a: The equilibria that exist when [Ru(h
6
-arene)(5,5’-difluorobipyridine)]
+
complexes are incubated in phosphate buffer. Figure 1b: Time course
19
F{
1
H} NMR
spectra of complex [4] incubated in 10mM deuterated phosphate buffer (2 mM Ru, pD =
7.2, 310 K). After 24 hr, 1 eq. of AgNO
3
was added to abstract the chloride ligand and
encourage formation of the aquo complex. Figure 1c:
19
F{
1
H} NMR spectra of complex [2]
incubated in D
2
O and buffered D
2
O of different phosphate concentration (2 mM Ru, pD =
7.2 (when buffered), 2 hr, 310 K). Figure 1d: Mass spectra recorded of the solution mixture
when complex [2] is incubated in D
2
O and buffered D
2
O, expected masses quoted in
Table S6.

5
With a number of titratable groups involved in these systems, examining the ligand
exchange behaviour of these complexes around conditions of pH relevant to biological
conditions requires strong buffering. We chose phosphate as a buffer also for its relevance
to biological conditions. However, it is clear that phosphate competes quite strongly as a
ligand to the metal , Figure 1c, and the presence of phosphate species have been
confirmed by mass spectrometry, Figure 1d and Table S6. Sadler et al. showed that the
structurally related [Ru(h
6
-arene)(en)Cl]
+
initially binds to the phosphate in the nucleobase
5-GMP before being displaced by the guanine N7.
41
To maintain a balance between
buffering strength and introducing a competing ligand a phosphate buffer concentration of
10 mM was chosen for all incubations, when the ruthenium concentration was 2 mM.
19
F resonances from the aquo and phosphate species are pH dependent and therefore the
pK
a
for the deprotonation of the bound aquo ligand can easily be measured. Figures 2a,
2b and 2c show representative data and calculated pK
a
values for the benzene complexes
with different bipyridyls (Complexes [1], [10] and [11]) and for the 5,5 difluorobipyridyl
complex with different arene ligands (Complexes [1] – [4]). The observed differences in
pK
a
are entirely consistent with the changes in electron density at the metal due to the
subtleties of the electron withdrawing effects of fluorines on the bipyridyl ligand and the
electron donating effects of the different arenes.
42
In other words, the trend observed
going from complex [1] – [4] shows that the increasing electron donating capability of the
h
6
-arene ligand to the ruthenium centre leads to a higher measured pK
a
value; an
increased electron density on the metal lowers its Lewis acidity. The electron withdrawing
capabilities of the trifluoromethyl group on complex [10] significantly increases the Lewis
acidity, reflected in the lowest pK
a
measured for any of these complexes. Interestingly,
pKa had to be measured using
1
H spectra because the
19
F resonances in this complex,
where the fluorine atoms are not directly on the bipyridyl ring, are not sensitive enough to
changes in coordination to accurately measure the pK
a
. Clearly this has implications for
the general use of
19
F on spectator ligands for reporting changes at the metal site.
![Figure 2a: pH dependence of the chemical shifts of the aquo coordinated ruthenium species and determination of the pKa in the following complexes [1] - Red circles; [2] - Blue squares; [3] - Pink triangles; [4] - black circles; [10] - Green squares; [11] - Orange triangles. 19F chemical shifts of the 5,5’-difluorobipyridyl fluorines or 1H chemical shift of the coordinated benzene ligand were plotted against pD. The lines are a least squares fit to an equation involving a single titratable group. Figure 2b: A series of 19F{1H} decoupled spectra of complex [1] in D2O phosphate buffer at differing pD values (2 mM Ru, 298 K). Figure 2c: The pKa values of a series of ruthenium complexes measured using (where applicable) both 19F{1H} NMR and 1H NMR. (a) – Unable to measure pKa from data collected. (b) – No fluorine atoms present in complex.](/figures/figure-2a-ph-dependence-of-the-chemical-shifts-of-the-aquo-1wfvdup5.png)
![Figure 6: A series of 19F{1H} NMR spectra when complex [1] is incubated with the protected amino acids, N-Ac-Cys-OMe, N-Z-Glu-OMe, N-Bz-His-OMe, N-Ac-Met-OMe, and a mixture of all amino acids together, (2 mM Ru, 3 eq. each amino acid, 24 hr, 310 K).](/figures/figure-6-a-series-of-19f-1h-nmr-spectra-when-complex-1-is-19oxlqsx.png)
![Figure 3: A 19F{1H} - 19F{1H} COSY spectra from the incubation of complex [1] with Nacetyl L-glutamine (2 mM Ru, 3 eq. amino acid, starting pD = 7.2, 24 hr, 310 K).](/figures/figure-3-a-19f-1h-19f-1h-cosy-spectra-from-the-incubation-of-1mpv5dl8.png)
![Figure 8: A series of 19F{1H} NMR spectra when complex [1] is incubated with defined mixtures of reduced and oxidised glutathione (2 mM Ru, 3 eq. glutathione, 24 hr, 310 K).](/figures/figure-8-a-series-of-19f-1h-nmr-spectra-when-complex-1-is-d1qj48xu.png)
![Figure 4: A series of 19F{1H} NMR spectra of complexes [1], [3], [5] and [7] incubated with N-acetyl cysteine methyl ester (2 mM Ru, 3 eq. amino acid, 310 K). The chemical shift scale is the same for all spectra, which are aligned to the chloride peak in each case. Absolute d values given in Table S5.](/figures/figure-4-a-series-of-19f-1h-nmr-spectra-of-complexes-1-3-5-2qcy121q.png)