




read more









To calculate collective degree, the authors used a total of 158, 102, and 202 ChIP-seq datasets in GM12878, H1-hESC, and K562 cell-types, respectively.
A significant depletion of motif instances at sites annotated by a label compared to other labels can very likely result in non-positive scores.
Most popular motif-finding methods use unsupervised machinelearning approaches to discover motifs in ‘foreground’ input sequences that are over-represented with respect to a set of ‘background’ sequences (e.g. “bound” vs. “unbound”, respectively) [1,2].
In other words, while the k-mer weight parameters for each subclass are learned directly from the data, the weight parameters for the labels are learned exclusively through the regularization constraint.
By implicitly accounting for the effects of overlapping annotation labels, SeqUnwinder can deconvolve sequence features associated with motor neuron programming dynamics and ES chromatin status.
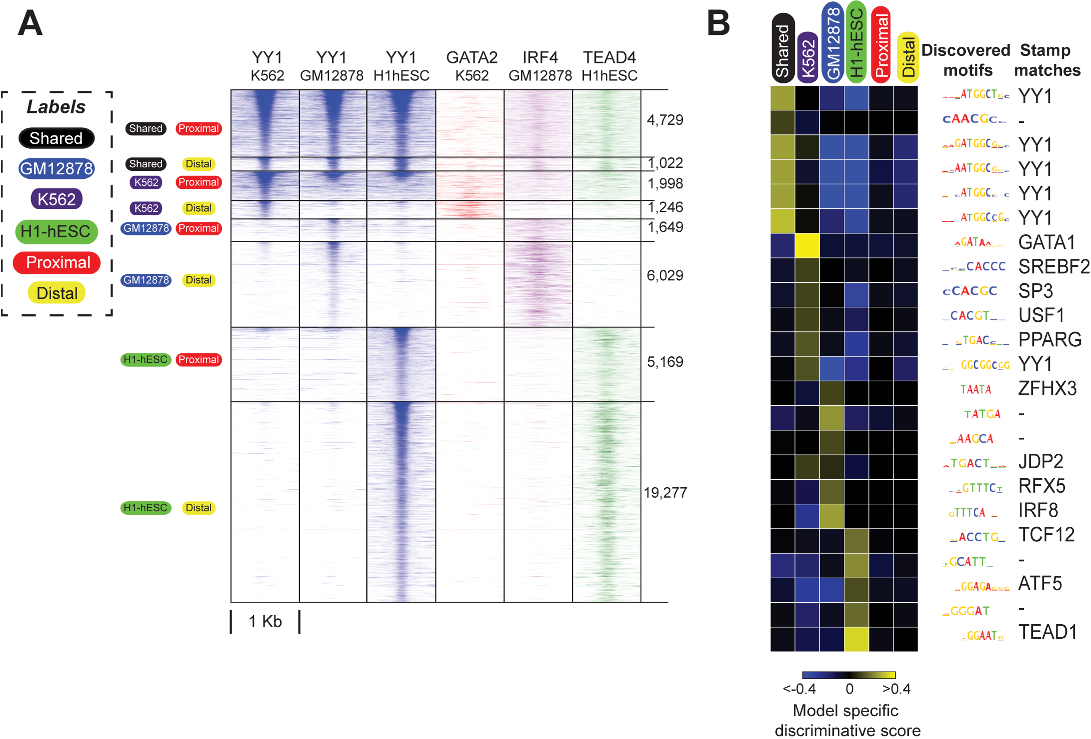
the authors found IRF and RUNX motifs enriched at GM12878-specific binding sites for 11 and 7 of the 17 examined TFs, respectively.
One advantage of the “hill-finding” approach is that it implicitly takes into account positional relationships between high-scoring k-mers on the genome; short stretches that contain multiple high-scoring k-mers will form larger “hills”.
Several variants of the basic string kernel (e.g. mismatch kernel [35], di-mismatch kernel [4], wild-card kernel [5,35], and gkm-kernel [36]) have been proposed and have been shown to substantially improve the classifier performance.
Since DREME takes only two classes as input: a foreground set and a background set, the authors ran four different DREME runs for each of the four labels.
binding sites showing significantly differential binding in any of the possible 3 pair-wise comparisons were removed from the shared set.
SeqUnwinder’s characterization of cell-specific motif features in collections of DNase-seq datasets may therefore serve as a source of predictive features for efforts that aim to predict cell-specific TF binding from accessibility experimental data alone [39–41].
To speed-up implementation, the authors restrict the unbiased k-mer features to only those k-mers that are present in at least 5% of the hills.
All sites with significantly greater Isl1/Lhx3 ChIP enrichment at 12h compared to 48h (q-value cutoff of<0.01) were labeled as “early”.
the motifs that the authors previously assigned to early or late TF binding behaviors could have been merely associated with ES-active and ES-inactive sites, respectively.
the cognate motif was not specifically predictive of cell-type-specific labels for the examined TFs, with the exception of H1-hESC-specific sites for CEBPB, NRSF and SRF.