Q1. What are the contributions in "Conserved motifs in the invertebrate iridescent virus 6 (iiv6) genome regulate virus transcription" ?
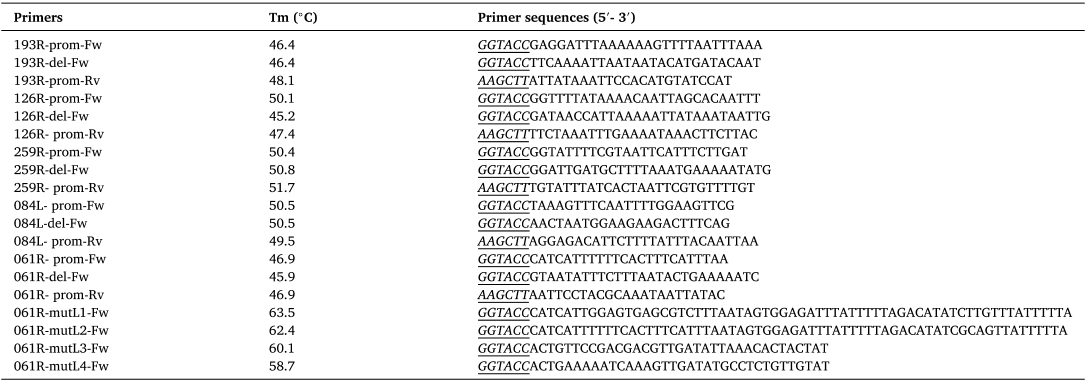
In this study, the authors investigated the transcriptional class of all IIV6 genes that had not been classified until now. Conversely, the presence of these two sequences upstream of the reporter decreased its expression. Next, upstream sequences of IIV6 L genes from which the authors removed this second motif in silico, were re-analyzed for the presence of potential conserved promoter sequences.