ARTICLE
Engineering designer beta cells with a CRISPR-Cas9
conjugation platform
Donghyun Lim
1,2,3
, Vedagopuram Sreekanth
1,2,3
, Kurt J. Cox
1,2,3
, Benjamin K. Law
1,2,3
, Bridget K. Wagner
1
,
Jeffrey M. Karp
4,5,6,7
& Amit Choudhary
1,2,3
✉
Genetically fusing protein domains to Cas9 has yielded several transformative technologies;
however, the genetic modifications are limited to natural polypeptide chains at the Cas9
termini, which excludes a diverse array of molecules useful for gene editing. Here, we report
chemical modifications that allow site-specific and multiple-site conjugation of a wide
assortment of molecules on both the termini and internal sites of Cas9, creating a platform
for endowing Cas9 with diverse functions. Using this platform, Cas9 can be modified to more
precisely incorporate exogenously supplied single-stranded oligonucleotide donor (ssODN)
at the DNA break site. We demonstrate that the multiple-site conjugation of ssODN to
Cas9 significantly increases the efficiency of precision genome editing, and such a platform is
compatible with ssODNs of diverse lengths. By leveraging the conjugation platform, we
successfully engineer INS-1E, a β-cell line, to repurpose the insulin secretion machinery,
which enables the glucose-dependent secretion of protective immunomodulatory factor
interleukin-10.
https://doi.org/10.1038/s41467-020-17725-0
OPEN
1
Chemical Biology and Therapeutics Science Program, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
2
Department of Medicine, Harvard
Medical School, Boston, MA 02115, USA.
3
Divisions of Renal Medicine and Engineering, Brigham and Women’s Hospital, Boston, MA 02115, USA.
4
Engineer ing in Medicine, Department of Medicine, Cent er for Regenerative Therapeutics, Brigham and Women’s Hospital, Harvard Medical School,
Boston, MA 02115, USA.
5
Harvar d− MIT Division of Health Sciences and Techno logy, MIT, Cambridge, MA 02139, USA.
6
Proteomics Platform, Broad
Institute of MIT and Harvard, Cambridge, MA 02142, USA.
7
Harvard Stem Cell Institute , Harvard University, Cambridge, MA 02138, USA.
✉
email: achoudhary@bwh.harva rd.edu
NATURE COMMUNICATIONS | (2020) 11:4043 | https://doi.org/10.1038/s41467-020-17725-0 | www.nature.com/naturecom munications 1
1234567890():,;

C
lustered regularly interspaced short palindromic repeats
(CRISPR)-Cas9 is a DNA endonuclease that can be tar-
geted to a genomic site using a guide RNA (gRNA)
bearing sequence complementarity to the target site
1
. The genetic
fusion of Cas9 with effector domains (e.g. a transcription acti-
vator) has yielded transformative technologies
2,3
; however, this
approach is limited to fusions that are generally linear, poly-
peptidic, and located on the termini of Cas9. A covalent con-
jugation platform that allows the creation of fusions that are non-
polypeptidic (e.g. nucleic acids, small molecules, polyethylene
glycol [PEG] chains), orthogonally branched from the internal
sites of Cas9, and amenable to multiple-site attachment to give a
multivalent display of functional cargos would provide a greater
diversity of technologies and applications. For example, precise
sequence alteration at the Cas9 cleavage site requires the efficient
incorporation of exogenously supplied single-stranded oligonu-
cleotide donor DNA (ssODN)
4
via the homology-directed repair
(HDR) pathway
5,6
. However, most cells instead employ the
non-homologous end-joining (NHEJ) repair, which results in
unpredictable insertions and deletions of bases at the cleavage
site, some of which are large enough to have pathogenic con-
sequences
7,8
. Displaying ssODNs on Cas9 can increase their local
concentration around the DNA strand break site to allow
enhanced incorporation of the desired sequence. In another
application, appending PEG chains to Cas9 may reduce the
immunogenicity of this bacterial protein
9
.
An ideal conjugation platform to Cas9 should have the fol-
lowing characteristics. First, the platform should be compatible
with a diverse set of cargos and allow their multiple-site attach-
ment. Second, the platform should be robust and implementable
by nonspecialists given the diverse users of CRISPR technologies.
Third, since some of the cargos (e.g. ssODN) are only available in
small quantities and are expensive, the conjugation system should
work efficiently without requiring large excesses. Ideally, the
platform should be modular and inexpensive to allow screening
of multiple conditions (e.g. ssODN sequence). Finally, for real-
world applications, the platform should allow scaled-up produc-
tion of the conjugates following good manufacturing practice
regulations
10
.
Herein, we present the development of such a platform that
relies on thiol-maleimide chemistry and DNA-base pairing,
which are both simple, well-established, scalable, and amenable to
a wide range of substrates
11
. We systematically scan the domains
of Cas9 to choose residues replaceable with engineered cysteines,
to which molecules of any size can be efficiently appended
without the loss of Cas9 activity. Because many possible con-
jugates (e.g. ssODNs) are prohibitively expensive for or una-
menable to direct thiol-maleimide conjugation, we also seek to
develop a more general conjugation platform. Thus, we design a
short oligonucleotide handle ‘adaptor’, which is attached to Cas9
via thiol-maleimide chemistry and uses base pairing to anchor
any molecule containing or appended with nucleic acids (Fig. 1a).
As an example of the platform’s utility, we use it to hybridize long
ssODNs that are large, expensive, and not available in sufficient
quantities for thiol-maleimide conjugation to Cas9. The resulting
Cas9:ssODN conjugates robustly enhance the precise incorpora-
tion of the desired sequence from ssODN in multiple cell types
and genomic sites. Importantly, the chemical conjugation plat-
form enables the multivalent display of ssODNs, which further
enhances the precise incorporation of the desired sequence over
that of the unitary display.
Next, we demonstrate the utility of our conjugation platform by
efficiently engineering insulin-producing β cells to secrete none-
ndogenous molecules, including an immunomodulatory protein
(∼160 residues), without incorporating any viral or foreign
sequences (e.g. promoter) other than that of the secreted molecule.
Current β-cell transplantation therapies for type 1 diabetes suffer
from immune rejection, resulting in acute cell loss and only short-
term therapeutic effects
12,13
. The macroencapsulation of β cells
with a semipermeable membrane can protect them from the host’s
immune system, although foreign body reaction-induced
fibrosis can impair the mass transfer and viability of encapsu-
lated cells
14–17
. Anti-inflammatory cytokines, such as interleukin
10 (IL-10), can reduce fibrosis and promote long-term β-cell
survival and superior islet function
18–20
. Therefore, engineered
β-cells that secrete anti-inflammatory cytokines and antifibrotic
factors can propel the development of cell-based therapeutics for
diabetes. Using our modified Cas9, we genetically repurpose the
insulin expression and secretion machinery to secrete a none-
ndogenous peptide and IL-10 in a glucose-responsive manner,
demonstrating an immediate usefulness of our genome editing
platform in developing cell-based therapeutics.
Results
Cas9 domains tolerate the attachment of diverse molecules.To
choose the sites for conjugation to Cas9, we analyzed the struc-
tures of apo-Cas9, gRNA-bound Cas9, and gRNA- and DNA-
bound Cas9 for residues that could provide a high labeling yield,
tolerate chemical modifications, span all the domains of Cas9,
and were surface-exposed in various Cas9 conformations for the
efficient modifications (Fig. 1b)
21–23
. Using the aforementioned
criteria, we identified two sites (204, 532) on the recognition
(REC) lobe, one site (826) on the HNH domain, five sites (1, 945,
1026, 1054, 1068) on the RuvC domain, and two sites (1153,
1207) on the protospacer adjacent motif-interacting (PI) domain.
We selected residues 558 and 1116 as controls, since modifica-
tions at 558 will impede the Cas9:gRNA interaction and at 1116
will impede PAM recognition by Cas9 (Fig. 1b and Supplemen-
tary Fig. 1). We optimized the conjugation conditions for Cas9
variants using biotin-maleimide and PEG (5 kDa)-maleimide as
model compounds to ensure that modifications of various sizes or
PI
HNH
RuvC
REC
BH
1207
1153
1116
826
1
1054
1068
945
1026
204
532
558
a
b
SH
SH
Cas9
Adaptor
Cas9
Cargo
Cargo
Cas9
Cas9
Cas9
Small molecule
Polymer
Fig. 1 A conjugation platform for Cas9. a A modular design strategy to
functionalize Cas9. b Structure-guided selection of chemical labeling sites.
Protospacer adjacent motif-interacting (PI) domain is in blue, HNH domain
is in yellow, RuvC domain is in cyan, recognition (REC) lobe is in magenta,
and bridge helix (BH) is in grey. Crystal structure of the Cas9-gRNA-DNA
ternary complex is used (PDB: 5F9R)
22
.
ARTICLE NATURE COMMUNICATIONS | https://doi.org/10.1038/s41467-020-17725-0
2 NATURE COMMUNICATIONS | (2020) 11:4043 | https://doi.org/10.1038/s41467-020-17725-0 | www.nature.com/naturecommunications
natures were tolerated (Supplementary Fig. 2). The reactions were
fast and high yielding at all sites except for the 1153 C mutant—
sites proximal to 1153 C (i.e. 1154 C) also yielded low conjugation
efficiencies (Supplementary Fig. 2). The location of these residues
was not assigned at the crystal structure of apo-Cas9 but the
residues were assumed to be amenable to efficient conjugation
since they were expected to be surface-exposed and flexible
21–23
.
Our labeling results, however, indicate that the loop may have
higher-order structures that prevent efficient chemical reactions,
so we did not use those sites in future experiments. To improve
the compatibility of the system with a broader range of con-
jugates, we next utilized the optimized reaction conditions to label
Cas9 at the remaining sites with a 17-nucleotide (nt) DNA
adaptor (5′-GCTTCACTCTCATCGTC-3′). The conversion rates
were comparable to those of PEG labeling (Supplementary Fig. 2),
demonstrating that the efficient conjugation of multiple cargo
types can occur at these sites. Thus, the identified sites provide
high conjugation yields with diverse molecules, including small
molecule and polymers (DNA or PEG).
To identify sites tolerant to the conjugation of the DNA
adaptor without the loss of Cas9 activity, we designed an ssODN
that would insert a 33-nt DNA fragment (HiBiT sequence
24
)at
the target gene (Supplementary Fig. 3). This insertion would
result in the expression of a fusion protein with a C-terminal
HiBiT tag, which is a small fragment of the NanoLuc luciferase.
When HiBiT is complemented by LgBiT, the remainder of
NanoLuc, the full-length luciferase is reconstituted to generate a
luminescence signal proportional to the degree of knockin,
providing an easy readout for HDR (Supplementary Fig. 3a). We
chose GAPDH as the first target gene (Supplementary Fig. 3b)
owing to its abundant expression in many cell types, which
should allow for the reliable detection of the luminescence signal.
Using the HiBiT knockin assay, we measured whether appending
the DNA adaptor to the cysteine affected Cas9 activity (Fig. 2a).
As expected, much of the Cas9 activity was lost by control
modifications at residues 558 and 1116, indicating the reliability
of the HiBiT knockin assay. We identified five sites whose activity
was largely maintained (>85% of wild-type in U2OS), even after
labeling with the 17-nt adaptor; these sites stemmed from three
regions: the REC lobe (532), the RuvC domain (1, 945, 1026), and
the PI domain (1207). To investigate the off-target profile of the
Cas9-adaptor conjugates, we used an eGFP disruption assay with
matched gRNA and mismatched gRNAs targeting the eGFP gene
of the U2OS.eGFP.PEST cells
25,26
. The Cas9-adaptor conjugate
retained the target specificity while also maintaining the on-target
activity (Supplementary Fig. 4a−c). Finally, we demonstrated that
Cas9s conjugated to the long PEG chain (5 kDa, Supplementary
Fig. 2c) retained the DNA cleavage activity in the eGFP
disruption assay, assuring that Cas9 could be modified with
diverse cargos without a loss of function (Supplementary Fig. 4d).
Unitary display of ssODN enhances HDR in several cell types.
Next, we designed ssODN with a sequence complementary to the
conjugation adaptor and confirmed the binding of the ssODN
bearing the complementary sequence to the Cas9-adaptor using a
gel-shift assay (Supplementary Fig. 5). To measure the ability of
ssODN conjugates to enhance HDR and the site dependence of such
enhancements, we performed the HiBiT knockin assay in U2OS
cells. Using the luminescence signals from unconjugated ssODN as
normalization controls, we observed enhanced knockin efficiency
at multiple sites (Fig. 2b and Supplementary Fig. 6a) with the
ssODN attached to Cas9. We were able to confirm such enhance-
ments in multiple cell lines, with a greater than four-fold increase in
HEK-293FT cells, around a 1.9-fold increase in human-induced
pluripotent stem cells, and a more than three-fold increase in pri-
mary fibr oblasts (Fig. 2c−f and Supplementary Fig. 6b−e). For
cells with higher HiBiT signal but lower HDR enhanceme nts, we
observed site dependence, with two internal conjugation sites (532,
945) generally performing better than the terminal conjugation
0.0
0.5
1.0
1.5
Relative luminescence
a
wt
1
204
532
558
826
945
1026
1054
1068
1116
1207
U2OS
U2OS
MDA-MB-231 HEK-293FT hiPSC
Primary
fibroblast
b
0.0
0.5
1.0
1.5
2.0
2.5
3.0
Fold knock-in enhancement
Fold knock-in enhancement
Fold knock-in enhancement
Fold knock-in enhancement
Fold knock-in enhancement
wt
1
532
945
1026
1207
f
0
1
2
3
4
5
6
wt
532
945
c
0
1
2
3
4
5
6
wt
1
532
945
1207
d
0
1
2
3
4
5
6
7
wt
1
532
945
1026
1207
e
0.0
0.5
1.0
1.5
2.0
2.5
3.0
wt
532
945
Fig. 2 Unitary display of ssODN on Cas9 domains enhances HDR in multiple cell types. a HiBiT knockin efficiencies by Cas9-adaptor conjugates
compared to unlabeled wild-type Cas9 (wt) when a separate Cas9/ssODN system was used (n = 3 biologically independent experiments except for 204
where n = 2). b–f ssODN display on Cas9 enhances HiBiT knockin efficiency in various cells: b U2OS, c MDA-MB-231, d HEK-293FT, e human-induced
pluripotent stem cells, and f primary human neonatal dermal fibroblasts. Unlabeled wild-type Cas9 (wt) and Cas9-adaptor conjugates labeled at the
indicated residues were used (n = 3 biologically independent experiments). Source data are provided as a Source Data file.
NATURE COMMUNICATIONS | https://doi.org/10.1038/s41467-020-17725-0 ARTICLE
NATURE COMMUNICATIONS | (2020) 11:4043 | https://doi.org/10.1038/s41467-020-17725-0 | www.nature.com/naturecommunications 3

site (1). An examination of the c rystal structure
22
indicates that
cargos on the two internal residues are expected to align with
substrate DNA, while cargos on the terminal residue project out-
ward from the DNA, which may explain the differences in the
HDR-enhancing capacities of different ssODN-bearing sites.
ssODN display platform allows rapid and facile screening.To
demonstrate the modular nature of our conjugation platform that
should allow the rapid testing of multiple conditions and to
confirm the generality of HDR enhancement by ssODN display,
we tested the ability of the conjugates to enhance HDR under
several scenarios (e.g. different genomic sites, ssODN sequences,
or readouts). Using the HiBiT knockin assay, we confirmed HDR
enhancements at another DNA cleavage site of the GAPDH locus
in U2OS (Fig. 3a and Supplementary Fig. 7a) and at multiple
genomic loci (PPIB in U2OS, CFL1 in HEK-293FT; Fig. 3a and
Supplementary Fig. 7b, c). We then demonstrated HDR
enhancement using a fluorescent readout and a longer knockin
fragment (GFP11, 57 nt). The correct incorporation of this
fragment generated detectable fluorescence through the expres-
sion of a fusion protein with a C-terminal GFP11 tag, which
forms a fully functional GFP when complemented by GFP1-10
(Supplementary Fig. 8)
27
. Here as well, displaying ssODNs on
Cas9 increased the knockin efficiency by more than three-fold
(Fig. 3b and Supplementary Fig. 7d). In addition to luminescence
and fluorescence readouts to demonstrate HDR enhancements,
we used a restriction endonuclease site knockin assay that
quantifies both NHEJ and HDR efficiencies at the CXCR4 locus
by gel electrophoresis (Supplementary Fig. 9), and observed the
increase in HDR efficiencies by more than two-fold when Cas9:
ssODN conjugates were employed (Fig. 3c). We then used a
previously reported droplet digital PCR (ddPCR) assay that
employs probes to distinguish between wild-type, NHEJ-edited,
and HDR-edited sequences at the RBM20 locus (Supplementary
Fig. 10a, b)
28,29
. All Cas9:ssODN conjugates increased the ratio of
HDR over NHEJ, again indicating the generality of our platform
(Fig. 3d). The conjugates also enhanced HDR when another
gRNA/ssODN pair was employed (Supplementary Fig. 10c).
Finally, we designed an assay to convert eGFP to BFP in U2OS
cells through HDR-based nucleotide exchange and found that
Cas9:ssODN conjugates enhanced precision genome editing for
this exogenous target gene as well (Supplementary Fig. 11).
Multivalent display of ssODN on Cas9 further enhances HDR.
Owing to the small size of our adaptor and the chemical nature of
our platform, multivalent displays are feasible (Fig. 4a). To
demonstrate multivalent display, we produced Cas9 double-
cysteine mutants (532 C/945 C and 532 C/1207 C) and attached
the adaptor to both sites (Supplementary Fig. 12a). Next, we
confirmed the binding of the ssODNs to Cas9 (Supplementary
Fig. 12b) and observed a boost in HDR efficiency for both the 33-
nt HiBiT insertion and the two-base exchange (Fig. 4b−d),
indicating that multivalent Cas9 internal modifications further
improve the functionality of conjugated Cas9 proteins. Finally, to
minimize the size of the labels on Cas9, we investigated the
possibility of further decreasing the length of the adaptor. To this
effect, we found that hybridization by 13 nt or 15 nt showed a
similar HDR-enhancing effect as the standard 17-nt pairing
(Supplementary Fig. 13).
Efficient engineering of β cells to secrete IL-10. To demonstrate
the functional applicability of our chemically modified Cas9, we
a
wt
532
wt
1
wt
532
wt
532
wt
532
wt
532
wt
945
wt
532
wt
945
wt
1026
wt
1207
0
1
2
3
Fold knock-in enhancement
Fold knock-in enhancement
0.0
0.5
1.0
1.5
2.0
2.5
0
1
2
3
4
5
GAPDH PPIB CFL1
HiBiT knock-in
0
1
2
3
4
5
b
GAPDH
GFP11 knock-in
CXCR4
12-base exchange
0.00
0.02
0.04
0.06
0.00
0.02
0.04
0.06
0.00
0.02
0.04
0.06
0.00
0.02
0.04
0.06
0.00
0.02
0.04
0.06
d
p = 0.072 p = 0.015 p = 0.053
p = 0.013 p = 0.035
RBM20
2-base exchange
0.0
0.1
0.2
0.3
HDR/NHEJ
HDR/NHEJ
0.0
0.1
0.2
0.3
c
p = 0.0069 p = 0.032
Fig. 3 ssODN display platform allows facile testing of multiple conditions. a HiBiT sequence knockin efficiency was increased at multiple genomic loci in
U2OS cells (GAPDH and PPIB) or HEK-293FT cells (CFL1), (n = 3 biologically independent experiments). b The GFP11 sequence insertion at the GAPDH
locus was promoted in HEK-293T cells (n = 3 biologically independent experiments). c HDR-mediated 12-base exchange efficiency at the CXCR4 locus was
increased in HEK-293T cells (n = 3 biologically independent experiments). d Two-base exchange at the RBM20 locus was promoted in HEK-293FT cells.
Unlabeled wild-type Cas9 (wt) and Cas9-adaptor conjugates labeled at the indicated residues were used (n = 3 biologically independent experiments).
P-values were calculated by paired two-tailed t-test. Source data are provided as a Source Data file.
ARTICLE NATURE COMMUNICATIONS | https://doi.org/10.1038/s41467-020-17725-0
4 NATURE COMMUNICATIONS | (2020) 11:4043 | https://doi.org/10.1038/s41467-020-17725-0 | www.nature.com/naturecommunications

used it to efficiently engineer β cells to endow the cells with
immunomodulatory function. Since C-peptide is cleaved during
proinsulin processing and is cosecreted with insulin, we hypo-
thesized that knocking in the desired gene into the C-peptide
portion of the proinsulin locus would enable the secretion of the
inserted gene product. Previously, a lentiviral vector encoding a
proinsulin-luciferase fusion construct, containing a luciferase
inserted into the C-peptide, expressed functional luciferase in
levels directly proportional to insulin when stably integrated into
the INS-1E rat β-cell line and responded sensitively to external
stimuli, such as glucose concentration
30
. However, viral-vector
engineering poses safety issues such as immunogenicity to viral
components or the unintended random insertion of DNA frag-
ments into the host genome
31,32
. Direct knockin of the desired
gene fragment into the C-peptide locus using Cas9 will allow
glucose-dependent cosecretion of the target gene products with
insulin. The knockin strategy does not require long regulatory
elements (e.g. promoters), which enables footprint-free and effi-
cient HDR because of the smaller knockin size. Any viral or
foreign sequences, which are often required to drive efficient gene
expression, can be avoided to minimize the immunogenicity
issues. In addition, due to the inherent high expression and
secretion level of insulin in β cells, it would be easy to modulate
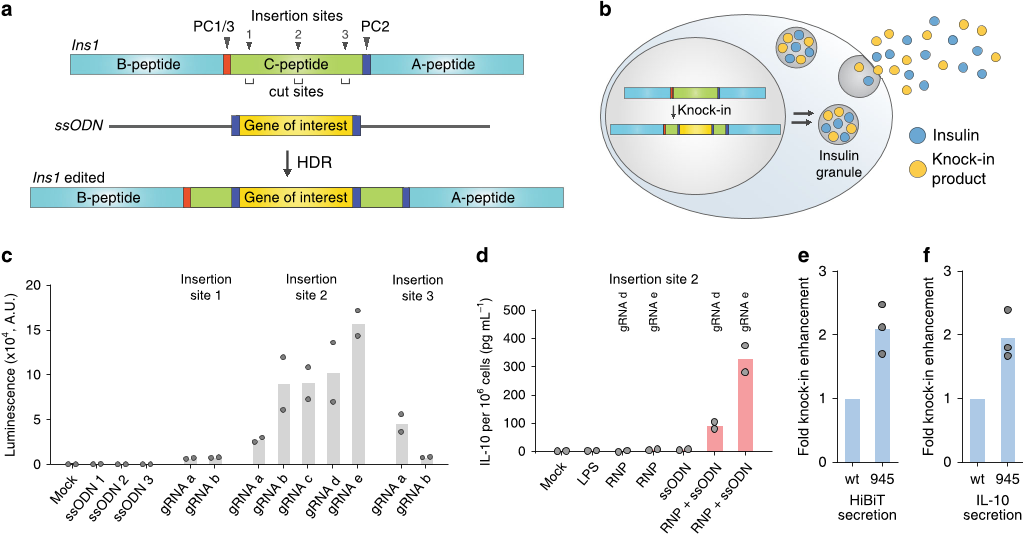
the secretion level of exogenous gene products (Fig. 5a, b).
We set out to demonstrate our β-cell engineering strategy with
the rat INS-1E β-cell line, which is widely used to study β-cell
biology. Murine cells have two insulin genes, Ins1 and Ins2, both
of which produce and secrete functional insulin; herein, we
targeted only the Ins1 gene. To identify the appropriate insertion
site in the Ins1 locus, we used HDR-mediated knockin of the
HiBiT sequence at the C-peptide portion (Fig. 5a). The target
HiBiT sequence was flanked by additional prohormone con-
vertase 2 (PC2) cleavage sites
30
to ensure no extra amino acids
would be present at each end of the knockin product after
processing (Fig. 5a). We chose three gene insertion sites at the
start, middle, and terminal regions of the C-peptide locus, and
designed several gRNAs to target these sites such that insertion
sites and DNA cleavage sites would be close enough to obtain
high HDR efficiency (Fig. 5a). In addition, genome-wide off-
target profiles of gRNAs were considered such that potential off-
target sites had mismatches at the seed sequences or at least three
mismatches in the gene-encoding regions. When standard
genome editing was performed at the target sites using
nonconjugated Cas9 and ssODN, the HiBiT peptide was secreted
from INS-1E cells, which could be readily detected through
luminescence signals from the cell culture supernatant after
complementation by the LgBiT protein. The highest knockin
efficiency was achieved by targeting the middle region of the C-
peptide (site 2) (Fig. 5c), so this insertion site was used for future
experiments. HiBiT peptide secretion was also stimulated by
glucose, consistent with the behavior expected from an insertion
at the ins1 locus (Supplementary Fig. 14a).
Based on this optimized design, we next knocked in Il10, whose
797-nt ssODN is much larger than that of HiBIT (183 nt). As our
approach leverages the insulin secretion pathway, the knockin
product will be secreted without the secretory signal peptide.
Thus, the signal peptide sequence present in the Il10 gene was
omitted when designing the knockin fragment. PC2 cleavage sites
were added at each end of Il10 to obtain intact IL-10 as the
knockin product, and the corresponding ssODN was synthesized
by reverse transcription. When INS-1E cells were transfected with
both unconjugated Cas9 and ssODN, IL-10 was secreted to the
cell culture medium as determined via enzyme-linked immuno-
sorbent assay (ELISA). No IL-10 was detected after transfection
with Cas9 or ssODN alone or in lipopolysaccharide (LPS)-treated
cells
33
(Fig. 5d). We confirmed the correct insertion of the Il10
gene at the Ins1 c-peptide region using Sanger sequencing
(Supplementary Fig. 15a−c). Finally, we conjugated ssODN on
Cas9 and found that both HiBiT secretion and IL-10 secretion
were significantly promoted by Cas9-ssODN conjugation over
that of separate Cas9 and ssODN (Fig. 5e, f and Supplementary
Fig. 16).
To further verify that the knockin products are released
through the regulated insulin secretion pathway, we investigated
the effect of insulin secretagogues
30
with distinct mode of actions.
To minimize the well-to-well signal differences originating from
the stochastic distribution of edited cells, we enriched the HiBiT
Cas9
SH
SH
ssODN
Adaptor
Cas9
a
b
532/945
0.0
0.5
1.0
1.5
2.0
2.5
3.0
Fold knock-in enhancement
0.0
0.5
1.0
1.5
2.0
2.5
3.0
p = 0.014
p = 0.0056
wt
532
532/945
wt
945
c
0.00
0.02
0.04
0.06
HDR/NHEJ
HDR/NHEJ
0.00
0.02
0.04
0.06
532/945
wt
532
532/945
wt
945
p = 0.028
p = 0.039
d
0.00
0.02
0.04
0.06
0.08
0.00
0.02
0.04
0.06
0.08
532/1207
wt
532
532/1207
wt
1207
p = 0.015
p = 0.039
Fig. 4 Multivalent display of ssODN further enhances HDR efficiency.
a Schematic illustrating the production of Cas9 double-ssODN conjugates.
b HiBiT sequence knockin at the GAPDH locus was detected in U2OS cells
(n = 4 biologically independent experiments). c, d Two-base exchange at
the RBM20 locus was detected in HEK-293FT cells. Unlabeled wild-type
Cas9 (wt) and Cas9-adaptor conjugates labeled at the indicated residues
were used (n = 3 biologically independent experiments). P-values were
calculated by paired two-tailed t -test. Source data are provided as a Source
Data file.
NATURE COMMUNICATIONS | https://doi.org/10.1038/s41467-020-17725-0 ARTICLE
NATURE COMMUNICATIONS | (2020) 11:4043 | https://doi.org/10.1038/s41467-020-17725-0 | www.nature.com/naturecommunications 5