




read more








The detection of further susceptibility loci will require genome-wide studies with more complete coverage and using larger numbers of cases and controls, together with the combination of results across multiple studies.
because recombination tends to occur at distinct ‘hot-spots’, neighbouring polymorphisms are often strongly correlated (in ‘linkage disequilibrium’, LD) with each other.
Known susceptibility genes account for less than 25% of the familial risk of breast cancer, and the residual genetic variance is likely to be due to variants conferring more moderate risks.
Most previously identified breast cancer susceptibility genes are involved in DNA repair, and many association studies in breast cancer have concentrated on genes in DNA repair and sex hormone synthesis and metabolism pathways.
Genotyping for stage 3, and for the fine-scale mapping of the FGFR2 locus, was conducted using either a 5′ nuclease assay (Taqman, Applied Biosystems) or MALDITOF mass spectrometry using the Sequenom iPLEX system.
Recent technological advances have provided platforms that allow hundreds of thousands of SNPs to be analysed in association studies, thus providing a basis for identifying moderate risk alleles without prior knowledge of position or function.
As most common cancers have similar familial relative risks to breast cancer, it is likely that similarly large studies will be required to identify common alleles for other cancers.
The cases were selected to have a strong family history of breast cancer, equivalent to at least two affected female first-degree relatives, because such cases are more likely to carry susceptibility alleles20.
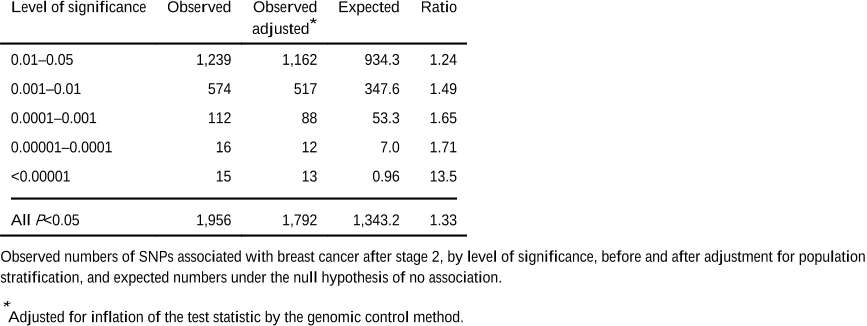
On the basis of their staged design and the estimated distribution of linkage disequilibrium between the typed SNPs and those in HapMap, the authors estimate that the power to identify the five most significant associations at P<10−7 (rs2981582, rs3803662, rs889312, rs13281615 and rs3817198) was 93%, 71%, 25%, 3% and 1% respectively.
the use of studies from multiple populations with different patterns of LD can substantially reduce the number of variants that need to be subjected to functional analysis.
It is notable that three of the five loci contain genes related to control of cell growth or to cell signalling, but only one (FGFR2) had a clear priorEaston et al.
In particular, three SNPs showed some evidence of association in stage 3 (P<0.05, in each case in the same direction as in stages 1Easton et al.